HighPlay: Cyclic Peptide Sequence Design Based on Reinforcement Learning and Protein Structure Prediction
IF 6.8
1区 医学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
The structural diversity and good biocompatibility of cyclic peptides have led to their emergence as potential therapeutic agents. Existing cyclic peptide design methods, whether traditional or emerging AI-assisted, rely on a multitude of experiments and face challenges such as limited molecular diversity, high cost, and time-consuming. In this study, we propose HighPlay, which integrates reinforcement learning (MCTS) with the HighFold structural prediction model to design cyclic peptide sequences based solely on the target protein sequence information, to achieve the synergistic optimization of cyclic peptide sequences and binding sites and to dynamically explore the sequence space without the need for predefined target information. The model was applied to the design of cyclic peptide sequences for three different targets, which were screened and verified through molecular dynamics simulations, demonstrating good binding affinity. Specifically, the cyclic peptide sequences designed for the TEAD4 target exhibited micromolar-level affinity in further experimental validation.
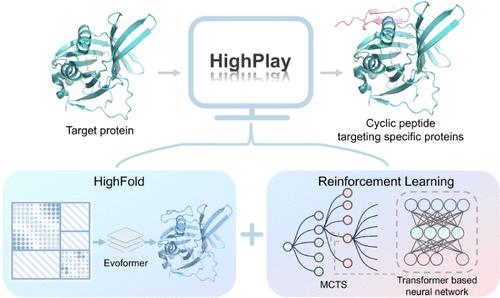
基于强化学习和蛋白质结构预测的循环肽序列设计
环肽结构的多样性和良好的生物相容性使其成为潜在的治疗药物。现有的环肽设计方法,无论是传统的还是新兴的人工智能辅助的,都依赖于大量的实验,面临着分子多样性有限、成本高、耗时等挑战。在本研究中,我们提出将强化学习(MCTS)与HighFold结构预测模型相结合的HighPlay,仅基于目标蛋白序列信息设计环肽序列,实现环肽序列和结合位点的协同优化,并在不需要预定义目标信息的情况下动态探索序列空间。将该模型应用于三种不同靶点的环肽序列设计,通过分子动力学模拟对其进行筛选和验证,显示出良好的结合亲和力。具体来说,为TEAD4靶点设计的环肽序列在进一步的实验验证中显示出微摩尔水平的亲和力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Medicinal Chemistry
医学-医药化学
CiteScore
4.00
自引率
11.00%
发文量
804
审稿时长
1.9 months
期刊介绍:
The Journal of Medicinal Chemistry is a prestigious biweekly peer-reviewed publication that focuses on the multifaceted field of medicinal chemistry. Since its inception in 1959 as the Journal of Medicinal and Pharmaceutical Chemistry, it has evolved to become a cornerstone in the dissemination of research findings related to the design, synthesis, and development of therapeutic agents.
The Journal of Medicinal Chemistry is recognized for its significant impact in the scientific community, as evidenced by its 2022 impact factor of 7.3. This metric reflects the journal's influence and the importance of its content in shaping the future of drug discovery and development. The journal serves as a vital resource for chemists, pharmacologists, and other researchers interested in the molecular mechanisms of drug action and the optimization of therapeutic compounds.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


