Tutorial: guidelines for the use of machine learning methods to mine genomes and proteomes for antibiotic discovery
IF 16
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
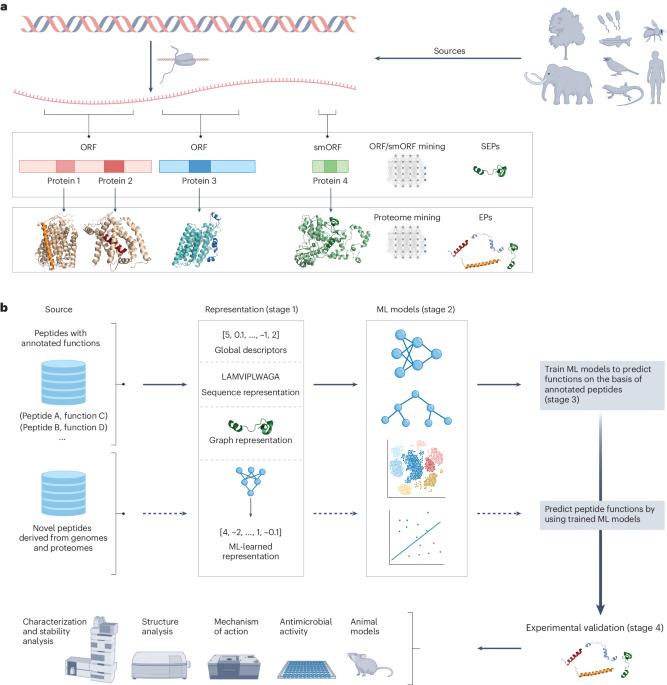
Genomes and proteomes constitute a rich reservoir of molecular diversity. However, they have remained underexplored because of a lack of appropriate tools. In recent years, computational approaches have been developed to mine this unexplored biological information, or dark matter, accelerating the discovery of new antibiotic molecules. Such efforts have yielded a wide range of new molecules. These include peptides released via predicted proteolytic cleavage of larger proteins, termed ‘encrypted peptides’, which have been found to be widespread in nature. Molecules encoded by and translated from small open reading frames within genomic sequences have also been uncovered, further expanding the landscape of bioactive compounds. Here, we discuss computational approaches, including machine learning and artificial intelligence (AI) tools, which have been used to date to identify antimicrobial compounds, with a special emphasis on peptides. We also propose potential avenues for future exploration in this rapidly evolving field. Moreover, we provide an overview of the experimental methods commonly used to validate these computational predictions. We anticipate that efforts combining cutting-edge AI and experimental approaches for biological sequence mining will reveal new insights into host immunity and continue to accelerate discoveries in the fields of antibiotics and infectious diseases. In this Tutorial, we provide guidelines for the use of ML models to mine genomes and proteomes for the discovery of antimicrobial peptides, discussing the various stages of the ML workflow, recent advances in ML models and how to experimentally validate the computational predictions.
教程:使用机器学习方法挖掘基因组和蛋白质组以发现抗生素的指南。
基因组和蛋白质组构成了丰富的分子多样性宝库。然而,由于缺乏适当的工具,它们仍未得到充分探索。近年来,计算方法已经发展到挖掘这些未被探索的生物信息,或暗物质,加速发现新的抗生素分子。这样的努力已经产生了大量的新分子。这些包括通过预测的较大蛋白质的蛋白水解裂解释放的肽,称为“加密肽”,已被发现在自然界中广泛存在。由基因组序列中的小开放阅读框编码和翻译的分子也被发现,进一步扩大了生物活性化合物的范围。在这里,我们讨论计算方法,包括机器学习和人工智能(AI)工具,迄今为止已用于识别抗菌化合物,特别强调肽。我们还提出了在这个快速发展的领域未来探索的潜在途径。此外,我们还概述了通常用于验证这些计算预测的实验方法。我们预计,结合尖端人工智能和生物序列挖掘实验方法的努力将揭示宿主免疫的新见解,并继续加速抗生素和传染病领域的发现。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


