Reclassification of variants of uncertain significance in neonatal genetic diseases: implications from a clinician’s perspective
IF 2.5
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
Although whole-exome sequencing (WES) is now widely used to diagnose neonatal genetic diseases, the genetic causes in over half of the cases remain unresolved, primarily due to variants of uncertain significance (VUS). Therefore, reclassifying VUS may be an effective strategy to improve WES’s diagnostic yield. However, not all reclassification approaches are suitable for clinicians. Patients in the neonatal unit of Hebei Provincial Children’s Hospital who underwent WES for suspected genetic diseases and demonstrated VUS were re-evaluated from January 2019 to December 2023 using user-friendly methods. A total of 676 individuals were tested, with 101 phenotype-associated VUS identified in 82 patients. Thirty (29.7%) VUS classifications were changed: 24 were upgraded to likely pathogenic or pathogenic, and 6 were downgraded to likely benign. VUS reclassification clarified the molecular diagnosis in 19 cases, increasing the WES diagnostic rate from 30.2% to 33.0%. Computational prediction contributed the most to reclassification, whereas clinical phenotype-related evidence was also particularly significant in upgrading variants. Moreover, phenotype-associated VUS with a score of ≥3 points are more likely to be classified as likely pathogenic or pathogenic, thus requiring more attention. This study provides a practical reference for clinicians in managing VUS reclassification.
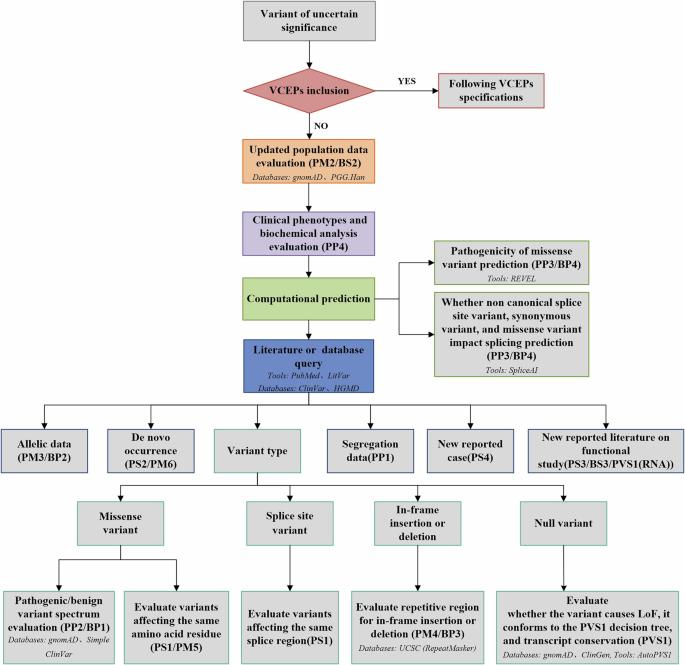
新生儿遗传疾病中不确定意义变异的重新分类:从临床医生角度的意义。
尽管全外显子组测序(WES)现在被广泛用于诊断新生儿遗传病,但超过一半的病例的遗传原因仍未得到解决,主要是由于不确定意义的变异(VUS)。因此,对VUS进行重新分类可能是提高WES诊断率的有效策略。然而,并不是所有的重新分类方法都适合临床医生。2019年1月至2023年12月,对河北省儿童医院新生儿病房因疑似遗传病接受WES检查并出现VUS的患者进行重新评估。总共对676人进行了测试,在82名患者中发现了101种与表型相关的VUS。30例(29.7%)VUS分类发生变化:24例升级为可能致病或致病,6例降级为可能良性。VUS重新分类澄清了19例的分子诊断,使WES诊断率由30.2%提高到33.0%。计算预测对重新分类贡献最大,而临床表型相关证据在升级变体方面也特别重要。此外,表型相关VUS评分≥3分更容易被归类为可能致病性或致病性,因此需要更多的关注。本研究为临床医生处理VUS重分提供了实用参考。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


