Group-specific Quantification of mcrA genes of Methanogenic Archaea and "Candidatus Methanoperedens" by Digital PCR.
IF 2
4区 环境科学与生态学
Q3 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Digital PCR is a technique that quantifies target genes based on the absence or presence of the targets in PCR amplicons. The present study exami-ned group-specific probes for the quantification of mcrA genes in six methanogenic archaeal groups and "Candidatus Methanoperedens" by digital PCR with the universal primers ML-f and ML-r. A digital PCR ana-lysis of paddy field soil detected all the targets, with the dominant and minor groups being Methanomicrobiales and Methanobrevibacter spp., respectively (107 and 104 copies [g dry soil]-1). This method has the potential to reveal the dynamics of specific methanogenic archaeal groups in the environment.
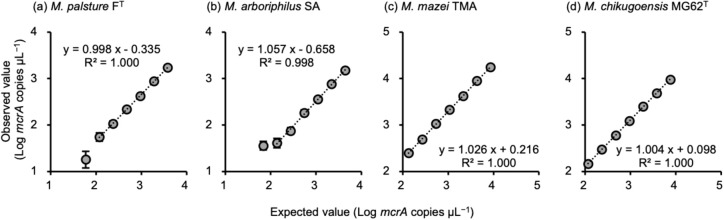
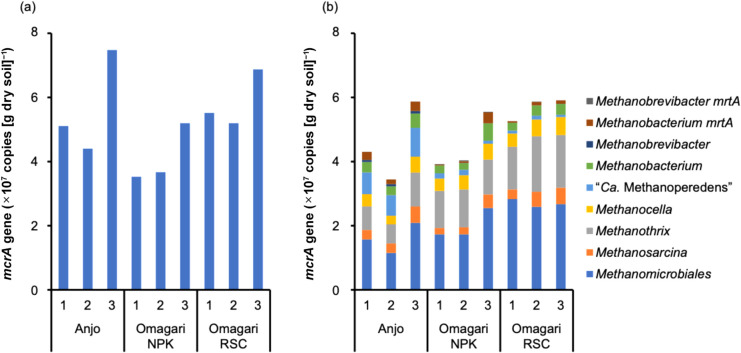
产甲烷古菌和“Methanoperedens Candidatus Methanoperedens”mcrA基因的群体特异性定量分析。
数字PCR是一种基于PCR扩增子中目标基因的缺失或存在来量化目标基因的技术。本研究以通用引物ML-f和ML-r为引物,对6个产甲烷古菌群和“Candidatus Methanoperedens”的mcrA基因进行了群体特异性PCR检测。对水田土壤进行数字PCR分析,检测到所有目标菌,优势群和次要群分别为Methanomicrobiales和Methanobrevibacter spp.(107和104拷贝[g干土]-1)。这种方法有可能揭示环境中特定产甲烷古菌群的动态。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Microbes and Environments
生物-生物工程与应用微生物
CiteScore
4.10
自引率
13.60%
发文量
66
审稿时长
3 months
期刊介绍:
Microbial ecology in natural and engineered environments; Microbial degradation of xenobiotic compounds; Microbial processes in biogeochemical cycles; Microbial interactions and signaling with animals and plants; Interactions among microorganisms; Microorganisms related to public health; Phylogenetic and functional diversity of microbial communities; Genomics, metagenomics, and bioinformatics for microbiology; Application of microorganisms to agriculture, fishery, and industry; Molecular biology and biochemistry related to environmental microbiology; Methodology in general and environmental microbiology; Interdisciplinary research areas for microbial ecology (e.g., Astrobiology, and Origins of Life); Taxonomic description of novel microorganisms with ecological perspective; Physiology and metabolisms of microorganisms; Evolution of genes and microorganisms; Genome report of microorganisms with ecological perspective.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


