SourceApp: A Novel Metagenomic Source Tracking Tool that can Distinguish between Fecal Microbiomes Using Genome-To-Source Associations Benchmarked Against Mixed Input Spike-In Mesocosms
IF 11.3
1区 环境科学与生态学
Q1 ENGINEERING, ENVIRONMENTAL
引用次数: 0
Abstract
Methodologies utilizing metagenomics are attractive to fecal source tracking (FST) aims for assessing the presence and proportions of various fecal inputs simultaneously. Yet, compared to established culture- or PCR-based techniques, metagenomic approaches for these purposes are rarely benchmarked or contextualized for practice. We performed shotgun sequencing experiments (n = 35) of mesocosms constructed from the water of a well-studied recreational and drinking water reservoir spiked with various fecal (n = 6 animal sources, 3 wastewater sources, and 1 septage source) and synthetic microbiome spike-ins (n = 1) introduced at predetermined cell concentrations to simulate fecal pollution events of known composition. We built source-associated genome databases using publicly available reference genomes and metagenome assembled genomes (MAGs) recovered from short- and long-read sequencing of the fecal spike-ins, and then created an associated bioinformatic tool, called SourceApp, for inferring source attribution and apportionment by mapping the metagenomic data to these genome databases. SourceApp’s performance varied substantially by source, with cows being underestimated due to under sampling of cow fecal microbiomes. Parameter tuning revealed sensitivity and specificity near 0.90 overall, which exceeded all alternative tools. SourceApp can assist researchers with analyzing and interpreting shotgun sequencing data and developing standard operating procedures on the frontiers of metagenomic FST.
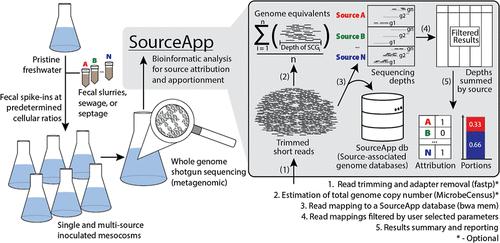
SourceApp:一种新的宏基因组源跟踪工具,可以使用基因组-源关联基准来区分粪便微生物组,以对抗混合输入峰值-中生态系统
利用宏基因组学的方法对于同时评估各种粪便输入的存在和比例的粪便源跟踪(FST)目标具有吸引力。然而,与已建立的基于培养或pcr的技术相比,用于这些目的的宏基因组方法很少有基准或实践背景。我们进行了鸟枪测序实验(n = 35),这些中生态系统是由一个经过充分研究的娱乐和饮用水水库的水组成的,其中加入了各种粪便(n = 6个动物源,3个废水源和1个污水源)和合成微生物群(n = 1),这些微生物群在预定的细胞浓度下加入,以模拟已知成分的粪便污染事件。我们利用公开的参考基因组和宏基因组组装基因组(MAGs)建立了来源相关的基因组数据库,这些基因组数据库是从粪便的短读和长读测序中恢复的,然后创建了一个相关的生物信息学工具,称为SourceApp,用于通过将宏基因组数据映射到这些基因组数据库来推断来源归因和分配。SourceApp的表现因来源而异,由于牛粪便微生物组取样不足,奶牛的表现被低估了。参数调整显示灵敏度和特异性接近0.90,超过所有替代工具。SourceApp可以帮助研究人员分析和解释霰弹枪测序数据,并在宏基因组FST领域开发标准操作程序。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

环境科学与技术
环境科学-工程:环境
CiteScore
17.50
自引率
9.60%
发文量
12359
审稿时长
2.8 months
期刊介绍:
Environmental Science & Technology (ES&T) is a co-sponsored academic and technical magazine by the Hubei Provincial Environmental Protection Bureau and the Hubei Provincial Academy of Environmental Sciences.
Environmental Science & Technology (ES&T) holds the status of Chinese core journals, scientific papers source journals of China, Chinese Science Citation Database source journals, and Chinese Academic Journal Comprehensive Evaluation Database source journals. This publication focuses on the academic field of environmental protection, featuring articles related to environmental protection and technical advancements.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


