A distinctive IGHV3-66 SARS-CoV-2 neutralizing antibody elicited by primary infection with an Omicron variant
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
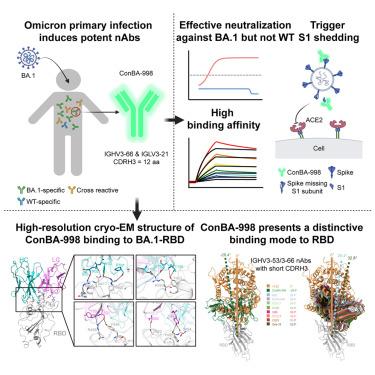
SARS-CoV-2 Omicron sub-variants continuously evolve under the pressure of neutralizing antibodies (nAbs), eliminating numerous potential elite monoclonal nAbs. The IGHV3-53/3-66 public nAbs have great potential for neutralizing SARS-CoV-2. However, it has been unclear whether a primary Omicron infection could also induce IGHV3-53/3-66 nAbs. In this study, we report an IGHV3-66-encoding monoclonal nAb, ConBA-998, that was elicited by primary infection with BA.1. ConBA-998 is an Omicron-dependent nAb with high binding affinity that triggers the shedding of the S1 subunit from the spike protein. The cryo-electron microscopy (cryo-EM) structure revealed the interactions between ConBA-998 and the Omicron BA.1 spike protein. ConBA-998 has a distinctive binding mode to receptor-binding domain (RBD) that differs from canonical IGHV3-53/3-66 nAbs. Overall, our findings indicate that Omicron may elicit unique specific nAbs distinct from those induced by pre-Omicron variants, providing further insights into SARS-CoV-2 variant-specific antibody responses.
一种独特的IGHV3-66 SARS-CoV-2中和抗体由原代感染引起
在中和抗体(neutralizing antibodies, nab)的压力下,SARS-CoV-2 Omicron亚变体不断进化,消除了许多潜在的精英单克隆nab。IGHV3-53/3-66公共nab具有中和SARS-CoV-2的巨大潜力。然而,尚不清楚原发性Omicron感染是否也能诱导IGHV3-53/3-66 nab。在这项研究中,我们报道了一种编码ighv3 -66的单克隆nAb, ConBA-998,它是由BA.1的原发性感染引发的。ConBA-998是一种高结合亲和力的omicron依赖性nAb,可触发刺突蛋白S1亚基的脱落。低温电镜(cryo-EM)结构揭示了ConBA-998与Omicron BA.1刺突蛋白之间的相互作用。ConBA-998与典型的IGHV3-53/3-66 nab具有不同的受体结合结构域(RBD)结合模式。总体而言,我们的研究结果表明,Omicron可能引发独特的特异性nab,不同于前Omicron变体诱导的nab,从而进一步了解SARS-CoV-2变体特异性抗体反应。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


