SagMSI: A graph convolutional network framework for precise spatial segmentation in mass spectrometry imaging
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
Mass Spectrometry Imaging (MSI) is a label-free imaging technique used in spatial metabolomics to explore the distribution of various metabolites within biological tissues. Spatial segmentation plays a crucial role in the biochemical interpretation of MSI data, yet the inherent complexity of the data—characterized by large size, high dimensionality, and spectral nonlinearity—poses significant analytical challenges in MSI segmentation. Although deep learning approaches based on convolutional neural networks (CNNs) have shown considerable success in spatial segmentation for biomedical imaging, they often struggle to capture the comprehensive structural information of MSI data.
Results
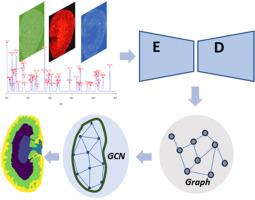
We propose SagMSI, an unsupervised graph convolution network (GCN)-based segmentation strategy that combines spatial-aware graph construction of MSI data with a GCN module within a deep neural network. This approach enables flexible, effective, and precise spatial segmentation. We applied SagMSI to both simulated data and various MSI experimental datasets and compared its performance against three commonly used segmentation methods, including t-SNE + k-means, a pipeline implemented by the R package Cardinal, and a CNN-based segmentation method. Visual comparisons with histological images and quantitative evaluations using the silhouette coefficient and adjusted rand index demonstrate that SagMSI excels in segmenting complex tissues, revealing detailed sub-structures, and delineating distinct boundaries of sub-organs with minimal noise interference. The integration of graph-based neural networks with spatially structural information offers deeper insights into spatial omics.
Significance
The MSI data is modelled by graph structure so as to incorporate the biomolecular profiling and spatial adjacency within neighboring pixels. The GCN framework generates meaningful pixel representations by learning local and global contextual information through the graph-based structure, thus enabling precise segmentation of MSI. The approach demonstrated high flexibility, robustness to noise, and applicability in exploring complex tissue structures and identifying marker ions associated with microregions.

质谱成像中精确空间分割的图卷积网络框架
质谱成像(MSI)是一种用于空间代谢组学的无标记成像技术,用于探索生物组织中各种代谢物的分布。空间分割在MSI数据的生化解释中起着至关重要的作用,然而数据的固有复杂性——大尺寸、高维和光谱非线性——给MSI数据分割带来了重大的分析挑战。尽管基于卷积神经网络(cnn)的深度学习方法在生物医学成像的空间分割方面取得了相当大的成功,但它们往往难以捕获MSI数据的全面结构信息。我们提出了一种基于无监督图卷积网络(GCN)的分割策略SagMSI,该策略将MSI数据的空间感知图构建与深度神经网络中的GCN模块相结合。这种方法可以实现灵活、有效和精确的空间分割。我们将SagMSI应用于模拟数据和各种MSI实验数据集,并将其与三种常用的分割方法(包括t-SNE+k-means,由R包Cardinal实现的管道和基于cnn的分割方法)的性能进行了比较。与组织学图像的视觉比较以及使用剪影系数和调整后的兰德指数进行的定量评估表明,SagMSI在分割复杂组织、揭示详细的亚结构和描绘亚器官的明显边界方面表现出色,且噪声干扰最小。基于图的神经网络与空间结构信息的集成为空间组学提供了更深入的见解。意义MSI数据采用图结构建模,结合了生物分子剖面和相邻像素间的空间邻接性。GCN框架通过基于图的结构学习局部和全局上下文信息,生成有意义的像素表示,从而实现对MSI的精确分割。该方法具有很高的灵活性,对噪声的鲁棒性,以及在探索复杂组织结构和识别与微区相关的标记离子方面的适用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


