Biodegradation of 1,2,4-trimethylbenzene in seawater using Rhodomonas sp. JZB-2: Performance, kinetics, and mechanism
IF 8.4
2区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
Abstract
Recently, marine ecosystems have been threatened by an accidental spill of C9 aromatics, particularly 1,2,4-trimethylbenzene (1,2,4-TMB), due to its high proportion in C9 aromatics. Microalgae-mediated bioremediation is a promising approach for pollutant removal owing to its eco-friendliness and carbon sequestration potential. In this study, the marine Cryptophyta Rhodomonas sp. JZB-2 demonstrated the ability to completely degrade 1–40 mg/L of 1,2,4-TMB within 6 days, showcasing its advantage in degrading 1,2,4-TMB at high concentrations compared to other microorganisms in the literature. Transcriptomics and proteomics analysis showed that several enzymes involved in 1,2,4-TMB degradation were significantly upregulated: hydroxylase (JmjC domain), iron/manganese-superoxide dismutase, and alcohol dehydrogenase etc. A new insight of biodegradation mechanism was elucidated that 1,2,4-TMB was initially oxidized by hydroxylase (JmjC domain) to 2,3,6-trimethylphenol, a process accelerated by the overexpression of iron/manganese-superoxide dismutase. Subsequently, 2,3,6-trimethylphenol was further degraded into 5-methylhexanoic acid via alcohol dehydrogenase and other short-chain dehydrogenases. Notably, the degradation products were less toxic than the parent compound (1,2,4-TMB). This study highlights the potential of Rhodomonas sp. JZB-2 for bioremediation of seawater contaminated with 1,2,4-TMB.
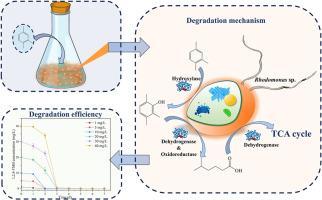
Rhodomonas sp. JZB-2降解海水中1,2,4-三甲基苯的性能、动力学及机理
近年来,由于C9芳烃中含有大量的1,2,4-三甲基苯(1,2,4- tmb),导致海洋生态系统受到严重威胁。微藻介导的生物修复由于其生态友好性和固碳潜力,是一种很有前途的污染物去除方法。在本研究中,海洋潜藻Rhodomonas sp. JZB-2能够在6天内完全降解1 - 40 mg/L的1,2,4- tmb,与文献中其他微生物相比,显示出其在高浓度下降解1,2,4- tmb的优势。转录组学和蛋白质组学分析显示,参与1,2,4- tmb降解的几个酶显著上调:羟化酶(JmjC结构域)、铁/锰超氧化物歧化酶和醇脱氢酶等。研究结果表明,1,2,4- tmb首先被羟化酶(JmjC结构域)氧化为2,3,6-三甲基苯酚,铁/锰超氧化物歧化酶的过表达加速了这一过程。随后,2,3,6-三甲基苯酚通过醇脱氢酶和其他短链脱氢酶进一步降解为5-甲基己酸。值得注意的是,降解产物的毒性低于母体化合物(1,2,4- tmb)。本研究强调了Rhodomonas sp. JZB-2对1,2,4- tmb污染海水的生物修复潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Environmental Management
环境科学-环境科学
CiteScore
13.70
自引率
5.70%
发文量
2477
审稿时长
84 days
期刊介绍:
The Journal of Environmental Management is a journal for the publication of peer reviewed, original research for all aspects of management and the managed use of the environment, both natural and man-made.Critical review articles are also welcome; submission of these is strongly encouraged.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


