Multimodal cell maps as a foundation for structural and functional genomics
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
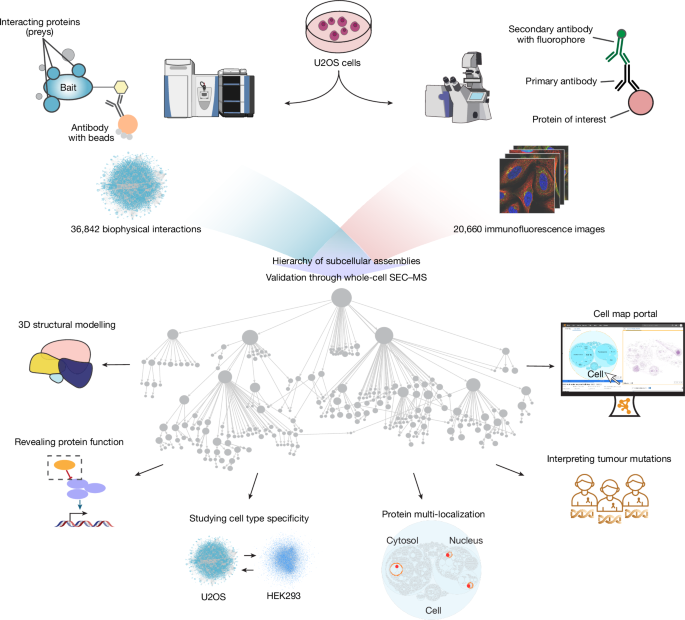
Human cells consist of a complex hierarchy of components, many of which remain unexplored1,2. Here we construct a global map of human subcellular architecture through joint measurement of biophysical interactions and immunofluorescence images for over 5,100 proteins in U2OS osteosarcoma cells. Self-supervised multimodal data integration resolves 275 molecular assemblies spanning the range of 10−8 to 10−5 m, which we validate systematically using whole-cell size-exclusion chromatography and annotate using large language models3. We explore key applications in structural biology, yielding structures for 111 heterodimeric complexes and an expanded Rag–Ragulator assembly. The map assigns unexpected functions to 975 proteins, including roles for C18orf21 in RNA processing and DPP9 in interferon signalling, and identifies assemblies with multiple localizations or cell type specificity. It decodes paediatric cancer genomes4, identifying 21 recurrently mutated assemblies and implicating 102 validated new cancer proteins. The associated Cell Visualization Portal and Mapping Toolkit provide a reference platform for structural and functional cell biology. A global map of human subcellular architecture yields protein complex structures, reveals protein functions, identifies assemblies with multiple localizations or cell-type specificity and decodes paediatric cancer genomes.

多模态细胞图谱是结构和功能基因组学的基础
人类细胞由复杂的层次结构组成,其中许多成分仍未被探索1,2。在这里,我们通过联合测量U2OS骨肉瘤细胞中超过5100种蛋白质的生物物理相互作用和免疫荧光图像,构建了人类亚细胞结构的全球地图。自监督多模态数据集成解决了275个分子组装,跨越10 - 8至10 - 5 m的范围,我们使用全细胞尺寸排除色谱法系统地验证并使用大型语言模型进行注释3。我们探索了结构生物学中的关键应用,产生了111个异二聚体复合物和一个扩展的rag - regulatory assembly的结构。该图谱为975种蛋白赋予了意想不到的功能,包括C18orf21在RNA加工中的作用和DPP9在干扰素信号传导中的作用,并鉴定了具有多种定位或细胞类型特异性的组装。它解码了儿童癌症基因组,鉴定了21个反复突变的组合,并暗示了102个经过验证的新癌症蛋白。相关的细胞可视化门户和绘图工具包为结构和功能细胞生物学提供了一个参考平台。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


