Shigella flexneri evades LPS ubiquitylation through IpaH1.4-mediated degradation of RNF213
IF 10.1
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
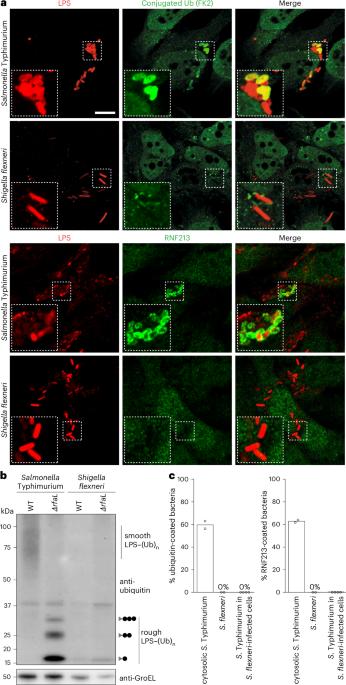
Pathogens have evolved diverse strategies to counteract host immunity. Ubiquitylation of lipopolysaccharide (LPS) on cytosol-invading bacteria by the E3 ligase RNF213 creates ‘eat me’ signals for antibacterial autophagy, but whether and how cytosol-adapted bacteria avoid LPS ubiquitylation remains poorly understood. Here, we show that the enterobacterium Shigella flexneri actively antagonizes LPS ubiquitylation through IpaH1.4, a secreted effector protein with ubiquitin E3 ligase activity. IpaH1.4 binds to RNF213, ubiquitylates it and targets it for proteasomal degradation, thus counteracting host-protective LPS ubiquitylation. To understand how IpaH1.4 recognizes RNF213, we determined the cryogenic electron microscopy structure of the IpaH1.4–RNF213 complex. The specificity of the interaction is achieved through the leucine-rich repeat of IpaH1.4, which binds the RING domain of RNF213 by hijacking the conserved RING interface required for binding to ubiquitin-charged E2 enzymes. IpaH1.4 also targets other E3 ligases involved in inflammation and immunity through binding to the E2-interacting face of their RING domains, including the E3 ligase LUBAC that is required for the synthesis of M1-linked ubiquitin chains on cytosol-invading bacteria downstream of RNF213. We conclude that IpaH1.4 has evolved to antagonize multiple antibacterial and proinflammatory host E3 ligases. Naydenova, Boyle and Pathe et al. report that Shigella uses the ubiquitin E3 ligase IpaH1.4 to evade lipopolysaccharide ubiquitylation in infected cells by degrading the host E3 ligase RNF213. Using cryo-electron microscopy, they present the structural basis of this interaction and the mechanism of immune evasion.

福氏志贺氏菌通过ipah1.4介导的RNF213降解来逃避LPS泛素化
病原体已经进化出多种对抗宿主免疫的策略。通过E3连接酶RNF213,入侵细胞质的细菌上脂多糖(LPS)的泛素化产生了抗菌自噬的“吃我”信号,但对于适应细胞质的细菌是否以及如何避免脂多糖泛素化仍然知之甚少。在这里,我们证明了福氏志贺氏肠杆菌通过IpaH1.4(一种具有泛素E3连接酶活性的分泌效应蛋白)有效地拮抗LPS泛素化。IpaH1.4结合RNF213,使其泛素化并靶向其蛋白酶体降解,从而抵消宿主保护LPS泛素化。为了了解IpaH1.4如何识别RNF213,我们确定了IpaH1.4 - RNF213复合物的低温电镜结构。这种相互作用的特异性是通过IpaH1.4富含亮氨酸的重复序列实现的,IpaH1.4通过劫持与带泛素的E2酶结合所需的保守RING界面,结合RNF213的RING结构域。IpaH1.4还通过结合其环结构域的e2相互作用面来靶向参与炎症和免疫的其他E3连接酶,包括在RNF213下游的细胞质侵入细菌上合成m1连接的泛素链所需的E3连接酶LUBAC。我们得出结论,IpaH1.4已经进化到可以拮抗多种抗菌和促炎宿主E3连接酶。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


