Profiling of runs of homozygosity from whole-genome sequence data in Japanese biobank
IF 2.5
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
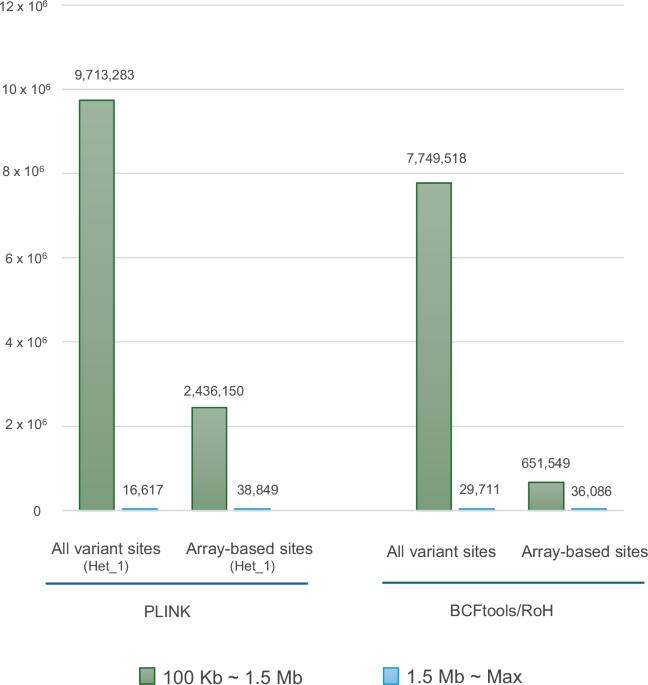
Runs of homozygosity (ROHs) are widely observed across the genomes of various species and have been reported to be associated with many traits and common diseases, as well as rare recessive diseases, in human populations. Although single nucleotide polymorphism (SNP) array data have been used in previous studies on ROHs, recent advances in whole-genome sequencing (WGS) technologies and the development of nationwide cohorts/biobanks are making high-density genomic data increasingly available, and it is consequently becoming more feasible to detect ROHs at higher resolution. In the study, we searched for ROHs in two high-coverage WGS datasets from 3552 Japanese individuals and 192 three-generation families (consisting of 1120 family members) in prospective genomic cohorts. The results showed that a considerable number of ROHs, especially short ones that may have remained undetected in conventionally used SNP-array data, can be detected in the WGS data. By filtering out sequencing errors and leveraging pedigree information, longer ROHs are more likely to be detected in WGS data than in SNP-array data. Additionally, we identified gene families within ROH islands that are associated with enriched pathways related to sensory perception of taste and odors, suggesting potential signatures of selection in these key genomic regions.
日本生物库全基因组序列数据的纯合性分析。
在各种物种的基因组中广泛观察到纯合子序列(ROHs),据报道,它与人类群体中的许多性状和常见疾病以及罕见隐性疾病有关。尽管单核苷酸多态性(SNP)阵列数据已被用于先前的ROHs研究,但近年来全基因组测序(WGS)技术的进步和全国队列/生物库的发展使得高密度基因组数据越来越多,因此以更高分辨率检测ROHs变得更加可行。在这项研究中,我们在两个高覆盖率的WGS数据集中搜索了ROHs,这些数据集来自3552名日本人和192个三代家庭(包括1120名家庭成员)的前瞻性基因组队列。结果表明,在WGS数据中可以检测到相当数量的ROHs,特别是在常规使用的snp阵列数据中可能无法检测到的短ROHs。通过过滤测序错误和利用谱系信息,在WGS数据中比在snp阵列数据中更容易检测到较长的ROHs。此外,我们在ROH岛屿中发现了与味觉和气味感官知觉相关的丰富通路相关的基因家族,这表明这些关键基因组区域存在潜在的选择特征。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


