Environmental sulfonamides pollution and microbial adaptation: Genome, transcriptome, and toxicology reveal Bacillus sp. HC-1 biotransformation and antibiotic resistance mechanisms
IF 11.3
1区 环境科学与生态学
Q1 ENGINEERING, ENVIRONMENTAL
引用次数: 0
Abstract
Sulfonamides (SAs) residue in the environment presents significant challenges to both environmental safety and medical security. Currently, the reaction and transformation mechanisms of microorganisms in the presence of SAs remain unclear. This study employed multiomics to investigate the gene response and enzymatic transformation mechanisms of Bacillus sp. HC-1 under SAs exposure conditions. Strain HC-1 demonstrated the ability to transform sulfaquinoxaline (SQX), sulfamethoxazole (SMX), and sulfamethazine (SMZ) into their respective N4-acetylated products. Within 12 hours, the transformation rates of SQX, SMX, and SMZ reached 51.7 %, 44.7 %, and 42.70 % respectively. Transcriptome analysis revealed that differentially expressed genes (DEGs) related to cellular transport, membrane channel activity, and various metabolic pathways were significantly enriched in strain HC-1 exposed to SQX. Through genomic analysis, we identified three types of arylamine N-acetyltransferases (NATs), which were named BaNATA, BaNATB, and BaNATC. Their highest homologies with reported NATs were 35.29 %, 40.82 %, and 35.32 %, respectively. Resistance and toxicological assessments indicated that NATs functioned as resistance genes against SAs, and the toxicity of transformation products to microorganisms and plant seeds was diminished. This study offers a valuable reference for a more in-depth understanding of microbial reactions, potential resistance, and transformation mechanisms in antibiotic-contaminated environments.
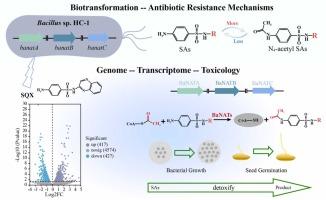
环境磺胺污染和微生物适应:基因组、转录组和毒理学揭示了芽孢杆菌HC-1的生物转化和抗生素耐药性机制
磺胺类药物在环境中的残留给环境安全和医疗保障带来了重大挑战。目前,微生物在sa存在下的反应和转化机制尚不清楚。本研究采用多组学方法研究了芽孢杆菌HC-1在sa暴露条件下的基因响应和酶转化机制。菌株HC-1表现出将磺胺喹啉(SQX)、磺胺甲恶唑(SMX)和磺胺甲基嗪(SMZ)转化为各自的n4乙酰化产物的能力。在12 h内,SQX、SMX和SMZ的转化率分别达到51.7%、44.7%和42.70%。转录组分析显示,暴露于SQX的HC-1菌株中,与细胞运输、膜通道活性和各种代谢途径相关的差异表达基因(DEGs)显著富集。通过基因组分析,我们鉴定出三种类型的芳胺n -乙酰转移酶(NATs),分别命名为BaNATA、BaNATB和BaNATC。与已报道的NATs的最高同源性分别为35.29%、40.82%和35.32%。抗性和毒理学评价表明,NATs具有抗性基因的功能,转化产物对微生物和植物种子的毒性减弱。本研究为深入了解抗生素污染环境中的微生物反应、潜在耐药性和转化机制提供了有价值的参考。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Hazardous Materials
工程技术-工程:环境
CiteScore
25.40
自引率
5.90%
发文量
3059
审稿时长
58 days
期刊介绍:
The Journal of Hazardous Materials serves as a global platform for promoting cutting-edge research in the field of Environmental Science and Engineering. Our publication features a wide range of articles, including full-length research papers, review articles, and perspectives, with the aim of enhancing our understanding of the dangers and risks associated with various materials concerning public health and the environment. It is important to note that the term "environmental contaminants" refers specifically to substances that pose hazardous effects through contamination, while excluding those that do not have such impacts on the environment or human health. Moreover, we emphasize the distinction between wastes and hazardous materials in order to provide further clarity on the scope of the journal. We have a keen interest in exploring specific compounds and microbial agents that have adverse effects on the environment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


