Amplification-Free Quantification of Endogenous Mitochondrial DNA Copy Number Using Solid-State Nanopores
IF 16
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
Mitochondrial DNA (mtDNA) quantification is crucial in understanding mitochondrial dysfunction, which is linked to a variety of diseases, including cancer and neurodegenerative disorders. Traditional methods often rely on amplification-based techniques, which can introduce bias and lack the precision needed for clinical diagnostics. Solid-state nanopores, an emerging biosensing platform, have the advantage of offering single-molecule and label-free approaches by enabling the direct counting of DNA molecules without amplification. The ion-current signatures obtained from each DNA molecule contain rich information on the molecules’ lengths and origin. In this study, we present an amplification-free method for mtDNA quantification using solid-state nanopores and machine learning. Intriguingly, we find that native (unamplified) mtDNA translocations harbor structurally distinctive features that can be exploited to specifically detect and quantify mtDNA copies over the background of genomic DNA fragments. By combining selective degradation of linear genomic DNA (gDNA) via exonuclease V with a support vector machine (SVM)-based model, we isolate and quantify mtDNA directly from biological samples. We validate our method using plasmids or isolated mtDNAs by spiking in predetermined quantities. We then quantify endogenous mtDNAs in a cancer cell line and in blood cells and compare our results with qPCR-based quantification of the mtDNA/nuclear DNA ratios. To elucidate the source of the ion-current signatures from the native mtDNA molecules, we perform synchronous electro-optical sensing of mtDNAs during passage through the nanopore after NHS ester reaction with fluorophore compounds. Our results show correlated electro-optical events, indicating that the mtDNA is complexed with packaging proteins. Our assay is robust, with a high classification accuracy and is capable of detecting mtDNA at picomolar levels, making it suitable for low-abundance samples. This technique requires minimal sample preparation and eliminates the need for amplification or purification steps. The developed approach has significant potential for point-of-care applications, offering a low-cost and scalable solution for accurate mtDNA quantification in clinical settings.
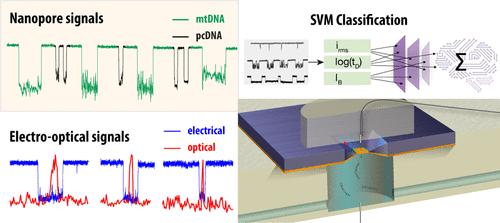
利用固态纳米孔进行内源线粒体DNA拷贝数的无扩增定量分析
线粒体DNA (mtDNA)定量对于理解线粒体功能障碍至关重要,线粒体功能障碍与多种疾病有关,包括癌症和神经退行性疾病。传统方法通常依赖于基于扩增的技术,这可能会引入偏差,并且缺乏临床诊断所需的精度。固态纳米孔是一种新兴的生物传感平台,其优势在于可以在不扩增的情况下直接计数DNA分子,从而提供单分子和无标记方法。从每个DNA分子中获得的离子电流特征包含有关分子长度和来源的丰富信息。在这项研究中,我们提出了一种使用固态纳米孔和机器学习的无扩增mtDNA定量方法。有趣的是,我们发现原生(未扩增的)mtDNA易位具有结构上的独特特征,可以用来在基因组DNA片段的背景下特异性地检测和量化mtDNA拷贝。通过结合外切酶V选择性降解线性基因组DNA (gDNA)和基于支持向量机(SVM)的模型,我们直接从生物样品中分离和量化了mtDNA。我们使用质粒或分离的mtdna通过预定数量的spike来验证我们的方法。然后,我们量化了癌细胞系和血细胞中的内源性mtDNA,并将我们的结果与基于qpcr的mtDNA/核DNA比率定量分析进行了比较。为了阐明天然mtDNA分子离子流特征的来源,我们在与荧光基团化合物发生NHS酯反应后通过纳米孔时对mtDNA进行同步电光传感。我们的结果显示相关的光电事件,表明mtDNA与包装蛋白络合。我们的分析是稳健的,具有很高的分类准确性,能够在皮摩尔水平检测mtDNA,使其适用于低丰度样品。该技术需要最少的样品制备,消除了扩增或纯化步骤的需要。开发的方法具有重大的护理点应用潜力,为临床环境中准确的mtDNA定量提供了低成本和可扩展的解决方案。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


