Context dependence in assembly code for supramolecular peptide materials and systems
IF 86.2
1区 材料科学
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
Abstract
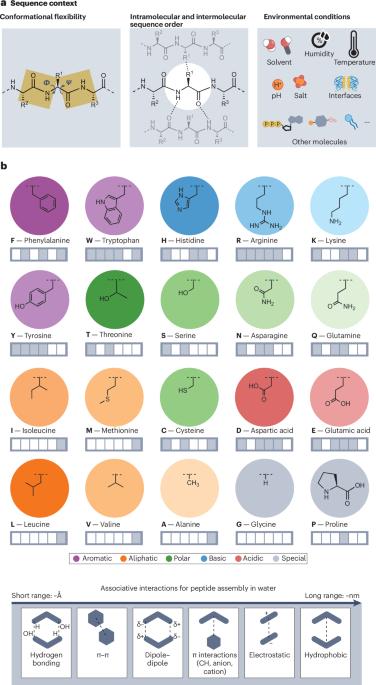
Living systems provide the most sophisticated materials known. These materials are created from a few dozen building blocks that are driven to self-organize by covalent and non-covalent interactions. Biology’s building blocks can be repurposed for the design of synthetic materials that life has not explored. In this Review, we examine the bottom-up design, discovery and evolution of self-assembling peptides by considering the entire supramolecular interaction space available to their constituent amino acids. Our approach focuses on sequence context, or how peptide sequence and environmental conditions collectively influence peptide self-assembly outcomes. We discuss examples of peptides that assemble through multimodal backbone, side chain and water interactions. We conclude that a more systematic (comparing sequences side-by-side), integrated (pairing computation and experiment) and holistic (considering peptide, solvent and environment) approach is required to better understand and fully exploit amino acids as a universal assembly code. This goal is particularly timely, because laboratory automation and artificial intelligence now have the potential to accelerate discoveries in these highly modular and complex materials, beyond the limited sequence space that biology uses. Living systems create exceptional materials from simple amino acid building blocks. This Review explores how a systems-based approach — considering peptide, solvent and environment, and integrating computation and experimentation — can unlock peptide sequence space as a universal materials assembly code, enabling designs beyond biology’s natural scope.

超分子肽材料和系统汇编代码中的上下文依赖性
生命系统提供了已知的最复杂的材料。这些材料是由几十个构建块创建的,这些构建块通过共价和非共价相互作用驱动自组织。生物学的构建模块可以重新用于设计生命尚未探索过的合成材料。在这篇综述中,我们研究了自组装肽的自下而上的设计,发现和进化,考虑到整个超分子相互作用空间可用于其组成氨基酸。我们的方法侧重于序列上下文,或肽序列和环境条件如何共同影响肽自组装结果。我们讨论了通过多模态主链、侧链和水相互作用组装的肽的例子。我们的结论是,需要一个更系统的(并排比较序列),集成的(配对计算和实验)和整体的(考虑肽,溶剂和环境)方法来更好地理解和充分利用氨基酸作为通用组装码。这一目标非常及时,因为实验室自动化和人工智能现在有可能加速这些高度模块化和复杂材料的发现,超越了生物学使用的有限序列空间。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Reviews Materials
Materials Science-Biomaterials
CiteScore
119.40
自引率
0.40%
发文量
107
期刊介绍:
Nature Reviews Materials is an online-only journal that is published weekly. It covers a wide range of scientific disciplines within materials science. The journal includes Reviews, Perspectives, and Comments.
Nature Reviews Materials focuses on various aspects of materials science, including the making, measuring, modelling, and manufacturing of materials. It examines the entire process of materials science, from laboratory discovery to the development of functional devices.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


