Integrating model systems and genomic insights to decipher mechanisms of cancer metastasis
IF 52
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
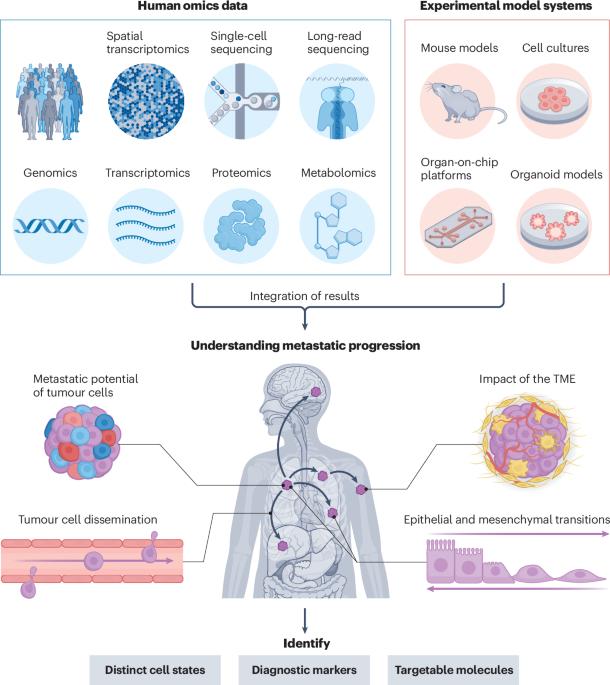
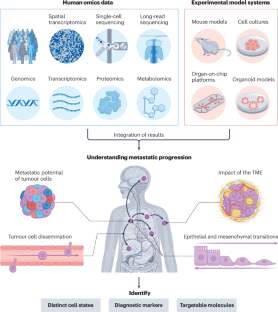
Deciphering metastatic processes is crucial for understanding cancer progression and potential treatment options. Genetic studies of model systems engineered to mimic metastatic disease, including organoids, genetically engineered mice and human cell lines, have had an important role in shaping our understanding of the metastatic cascade and how it can be manipulated. More recently, advances in high-throughput sequencing have enabled human metastases to be studied at single-cell and single-nucleotide resolution, providing insights into metastatic evolution and phenotypes of both cancer cells and immune cells. However, human tissue studies are often correlative and descriptive, whereas experimental models are reductionistic by nature, meaning that individual results should be interpreted with caution. Crucially, these seemingly disparate branches of metastasis research can and should complement each other to strengthen and validate findings. Here we explore the synergies between model systems and sequencing studies and outline key areas that must be explored to improve our understanding of the metastatic process. This Review explores how experimental models of metastasis, such as mouse models and cell cultures, can complement the (multi)omics analysis of human metastasis samples, thereby filling knowledge gaps left by model studies and validating the findings from human sequencing data.
整合模型系统和基因组的见解来破译癌症转移的机制
破译转移过程对于了解癌症进展和潜在的治疗方案至关重要。对模拟转移性疾病(包括类器官、基因工程小鼠和人类细胞系)的模型系统的遗传研究,在塑造我们对转移级联及其如何被操纵的理解方面发挥了重要作用。最近,高通量测序技术的进步使人类转移瘤能够在单细胞和单核苷酸分辨率下进行研究,为癌细胞和免疫细胞的转移进化和表型提供了见解。然而,人体组织研究通常是相关的和描述性的,而实验模型本质上是简化的,这意味着个体结果应该谨慎解释。至关重要的是,这些看似不同的转移研究分支可以而且应该相互补充,以加强和验证研究结果。在这里,我们探讨了模型系统和测序研究之间的协同作用,并概述了必须探索的关键领域,以提高我们对转移过程的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


