Single-cell lipidomics: protocol development for reliable cellular profiling using capillary sampling
IF 3.6
3区 化学
Q2 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
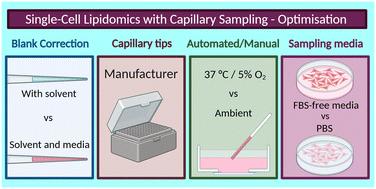
Single-cell lipidomics enables detailed analysis of the lipidomes of cells, but is challenged by small sample volumes, the risk of background interference and a lack of validation data. In this study, we explore the effect of different sampling variables on the lipid profiles of single pancreatic cancer cells, detected using liquid chromatography-mass spectrometry (LC-MS). We use automated and manual capillary sampling methods to isolate living single cells and evaluate different sampling media, capillary tips, aspiration volume, and temperature and humidity control. We demonstrate that automated and manual capillary sampling yield comparable lipid profiles when key parameters are controlled. Our findings highlight that appropriate blank correction, capillary tip type, and the control of aspiration volumes are all critical to preserving detection sensitivity. Conversely, choice of sampling medium does not affect lipidomics results. We also set out suggested best practices for these methodological variables, laying a foundation for robust, adaptable workflows in single-cell lipidomics for applications such as biomarker discovery and metabolic research.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analyst
化学-分析化学
CiteScore
7.80
自引率
4.80%
发文量
636
审稿时长
1.9 months
期刊介绍:
"Analyst" journal is the home of premier fundamental discoveries, inventions and applications in the analytical and bioanalytical sciences.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


