Highly Reusable Enzyme-Driven DNA Logic Circuits
IF 15.8
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
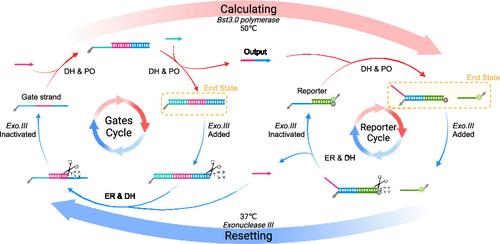
In recent years, DNA has emerged as a promising molecule for the construction of molecular computing systems. In the research field of DNA logic circuits, enzyme-driven DNA logic circuits, which offer faster reactions and lower complexity, have become the key focus in the field. However, it remains a significant drawback that it lacks the capability of being reused. Reusability is essential to enhance the computational capacity, correct errors, and reduce costs in DNA circuits. In this study, we propose a method for achieving high reuse in enzyme-driven DNA logic circuits using exonuclease III. By selectively digesting ds-DNA while preserving gate strands, our system highly restores the circuit to its initial state, which contains no waste-strand. This reuse method has demonstrated good performance in the converted-input reuse experiment of single-gate, multilayer cascades. Finally, we achieve four times converted-input reuse in a relatively complex circuit and three times multiple reuse in a square root DNA circuit.
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


