Rapid brain tumor classification from sparse epigenomic data
IF 58.7
1区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
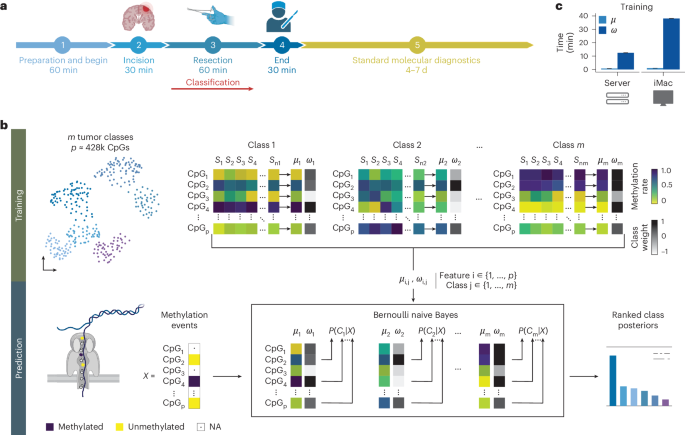
Although the intraoperative molecular diagnosis of the approximately 100 known brain tumor entities described to date has been a goal of neuropathology for the past decade, achieving this within a clinically relevant timeframe of under 1 h after biopsy collection remains elusive. Advances in third-generation sequencing have brought this goal closer, but established machine learning techniques rely on computationally intensive methods, making them impractical for live diagnostic workflows in clinical applications. Here we present MethyLYZR, a naive Bayesian framework enabling fully tractable, live classification of cancer epigenomes. For evaluation, we used nanopore sequencing to classify over 200 brain tumor samples, including 10 sequenced in a clinical setting next to the operating room, achieving highly accurate results within 15 min of sequencing. MethyLYZR can be run in parallel with an ongoing nanopore experiment with negligible computational overhead. Therefore, the only limiting factors for even faster time to results are DNA extraction time and the nanopore sequencer’s maximum parallel throughput. Although more evidence from prospective studies is needed, our study suggests the potential applicability of MethyLYZR for live molecular classification of nervous system malignancies using nanopore sequencing not only for the neurosurgical intraoperative use case but also for other oncologic indications and the classification of tumors from cell-free DNA in liquid biopsies. MethyLYZR, an epigenetic classifier of brain tumors, provides clinically relevant cancer classification results within 15 min of sequencing, with potential applications for neurosurgical intraoperative use.

从稀疏的表观基因组数据快速分类脑肿瘤
尽管在过去的十年中,对大约100种已知脑肿瘤实体的术中分子诊断一直是神经病理学的目标,但在活检收集后不到1小时的临床相关时间内实现这一目标仍然是难以捉摸的。第三代测序技术的进步使这一目标更加接近,但现有的机器学习技术依赖于计算密集型的方法,这使得它们在临床应用中的实时诊断工作流程中不切实际。在这里,我们提出了MethyLYZR,这是一个朴素的贝叶斯框架,可以对癌症表观基因组进行完全可处理的实时分类。为了评估,我们使用纳米孔测序对200多个脑肿瘤样本进行了分类,其中10个样本在手术室旁边的临床环境中进行了测序,在测序后15分钟内获得了高度准确的结果。MethyLYZR可以与正在进行的纳米孔实验并行运行,计算开销可以忽略不计。因此,更快得到结果的唯一限制因素是DNA提取时间和纳米孔测序仪的最大并行吞吐量。虽然需要更多的前瞻性研究证据,但我们的研究表明,MethyLYZR在神经系统恶性肿瘤的活体分子分类中具有潜在的适用性,不仅适用于神经外科术中应用,也适用于其他肿瘤适应症和液体活检中无细胞DNA的肿瘤分类。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Medicine
医学-生化与分子生物学
CiteScore
100.90
自引率
0.70%
发文量
525
审稿时长
1 months
期刊介绍:
Nature Medicine is a monthly journal publishing original peer-reviewed research in all areas of medicine. The publication focuses on originality, timeliness, interdisciplinary interest, and the impact on improving human health. In addition to research articles, Nature Medicine also publishes commissioned content such as News, Reviews, and Perspectives. This content aims to provide context for the latest advances in translational and clinical research, reaching a wide audience of M.D. and Ph.D. readers. All editorial decisions for the journal are made by a team of full-time professional editors.
Nature Medicine consider all types of clinical research, including:
-Case-reports and small case series
-Clinical trials, whether phase 1, 2, 3 or 4
-Observational studies
-Meta-analyses
-Biomarker studies
-Public and global health studies
Nature Medicine is also committed to facilitating communication between translational and clinical researchers. As such, we consider “hybrid” studies with preclinical and translational findings reported alongside data from clinical studies.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


