Deactivated Cas9-Engineered Magnetic Micromotors toward a Point-of-Care Digital Viral RNA Assay
IF 15.8
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
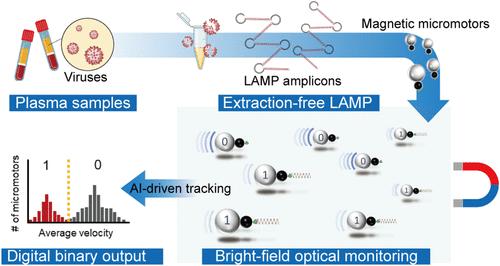
Digital nucleic acid assays, known for their high sensitivity and specificity, typically rely on fluorescent readouts and expensive and complex nanowell manufacturing, which constrain their broader use in point-of-care (POC) application. Here, we introduce an alternative digital molecular diagnostics, termed dCRISTOR, by seamlessly integrating deactivated Cas9 (dCas9)-engineered micromotors, extraction-free loop-mediated isothermal amplification (LAMP), low-cost bright field microscopy, and deep learning-enabled image processing. The micromotor, composed of a polystyrene sphere attached to a magnetic bead, incorporates a dCas9 ribonucleoprotein complex. The presence of human immunodeficiency virus-1 (HIV-1) RNA in a sample results in the formation of large-sized amplicons that can be specifically captured by the micromotors, reducing their velocity induced by an external magnetic field. The micromotor is propelled by an external magnetic field, which eliminates the need for chemical fuels, reducing system complexity, and allowing for precise control over micromotor movement, enhancing accuracy and reliability. A convolutional neural network classification-based multiobject tracking algorithm, CNN-MOT, accurately measures the change in micromotor motion, facilitating the binary digital assay format (“1” or “0”) for simplified result interpretation without user bias. Incorporating an extraction-free LAMP assay streamlines the dCRISTOR workflow, enabling qualitative HIV-1 detection in spiked plasma (n = 21) that demonstrates 100% sensitivity and specificity and achieves a limit of detection (LOD) of 0.96 copies/μL. The assay also achieved 100% correlation with reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in clinical patient samples (n = 9). The dCRISTOR assay, a label-free digital nucleic acid testing system that eliminates the need for fluorescence readouts, absorbance measurements, or expensive manufacturing processes, represents a substantial advancement in digital viral RNA diagnostics.
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


