Neutrophil recruitment during intestinal inflammation primes Salmonella elimination by commensal E. coli in a context-dependent manner
IF 20.6
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
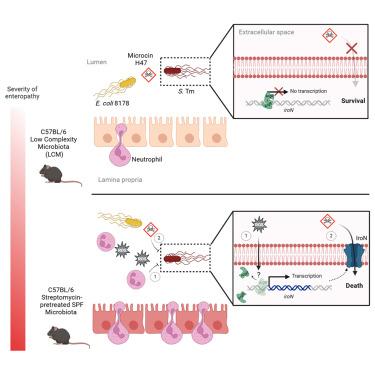
Foodborne bacterial diarrhea involves complex pathogen-microbiota-host interactions. Pathogen-displacing probiotics are increasingly popular, but heterogeneous patient outcomes highlighted the need to understand individualized host-probiotic activity. Using the mouse gut commensal Escherichia coli 8178 and the human probiotic E. coli Nissle 1917, we found that the degree of protection against the enteric pathogen Salmonella enterica serovar Typhimurium (S. Tm) varies across mice with distinct gut microbiotas. Pathogen clearance is linked to enteropathy severity and subsequent recruitment of intraluminal neutrophils, which differs in a microbiota-dependent manner. By combining mouse knockout and antibody-mediated depletion models with bacterial genetics, we show that neutrophils and host-derived reactive oxygen species directly influence E. coli-mediated S. Tm displacement by potentiating siderophore-bound toxin killing. Our work demonstrates how host immune factors shape pathogen-displacing probiotic efficiency while also revealing an unconventional antagonistic interaction where a gut commensal and the host synergize to displace an enteric pathogen.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


