Antibiotic target discovery by integrated phenotypic and activity-based profiling of electrophilic fragments
IF 6.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The emergence of antibiotic resistance necessitates the discovery of novel bacterial targets and antimicrobial agents. Here, we present a bacterial target discovery framework that integrates phenotypic screening of cysteine-reactive fragments with competitive activity-based protein profiling to map and functionally characterize the targets of screening hits. Using this approach, we identify β-ketoacyl-acyl carrier protein synthase III (FabH) and MiaA tRNA prenyltransferase as primary targets of a hit fragment, 10-F05, that confer bacterial stress resistance and virulence in Shigella flexneri. Mechanistic investigations elucidate that covalent C112 modification in FabH, an enzyme involved in bacterial fatty acid synthesis, results in its inactivation and consequent growth inhibition. We further demonstrate that irreversible C273 modification at the MiaA RNA-protein interaction interface abrogates substrate tRNA binding, attenuating resistance and virulence through decreased translational accuracy. Our findings underscore the efficacy of integrating phenotypic and activity-based profiling of electrophilic fragments to accelerate the identification and pharmacologic validation of new therapeutic targets.
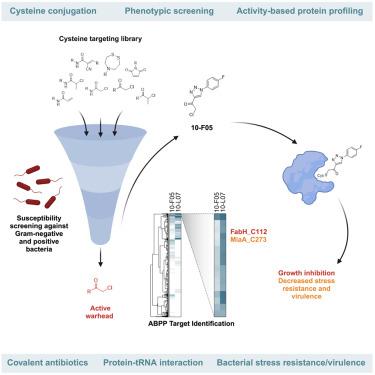

通过亲电片段的综合表型和活性分析发现抗生素靶标
抗生素耐药性的出现要求发现新的细菌靶点和抗菌剂。在这里,我们提出了一个细菌靶点发现框架,该框架将半胱氨酸反应片段的表型筛选与基于竞争活性的蛋白质分析相结合,以绘制和功能表征筛选靶点。利用这种方法,我们确定β-酮酰基-酰基载体蛋白合成酶III (FabH)和MiaA tRNA戊烯基转移酶是一个命中片段10-F05的主要靶标,该片段赋予了福氏志贺氏菌的细菌抗性和毒力。机制研究表明,参与细菌脂肪酸合成的酶FabH的共价C112修饰导致其失活并随之抑制生长。我们进一步证明,在MiaA rna -蛋白质相互作用界面上不可逆的C273修饰消除了底物tRNA的结合,通过降低翻译准确性来减弱抗性和毒力。我们的研究结果强调了整合亲电片段的表型和基于活性的分析来加速新治疗靶点的识别和药理学验证的有效性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell Chemical Biology
Biochemistry, Genetics and Molecular Biology-Molecular Medicine
CiteScore
14.70
自引率
2.30%
发文量
143
期刊介绍:
Cell Chemical Biology, a Cell Press journal established in 1994 as Chemistry & Biology, focuses on publishing crucial advances in chemical biology research with broad appeal to our diverse community, spanning basic scientists to clinicians. Pioneering investigations at the chemistry-biology interface, the journal fosters collaboration between these disciplines. We encourage submissions providing significant conceptual advancements of broad interest across chemical, biological, clinical, and related fields. Particularly sought are articles utilizing chemical tools to perturb, visualize, and measure biological systems, offering unique insights into molecular mechanisms, disease biology, and therapeutics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


