Autophagy repression by antigen and cytokines shapes mitochondrial, migration and effector machinery in CD8 T cells
IF 27.7
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
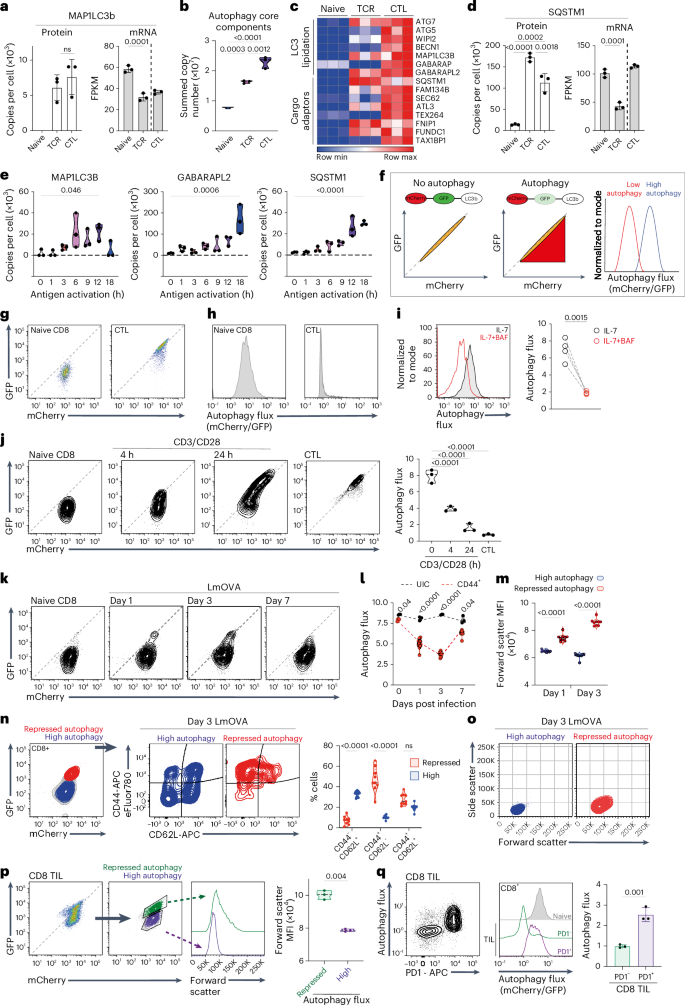
Autophagy shapes CD8 T cell fate; yet the timing, triggers and targets of this process are poorly defined. Herein, we show that naive CD8 T cells have high autophagic flux, and we identify an autophagy checkpoint whereby antigen receptor engagement and inflammatory cytokines acutely repress autophagy by regulating amino acid transporter expression and intracellular amino acid delivery. Activated T cells with high levels of amino acid transporters have low autophagic flux in amino-acid-replete conditions but rapidly reinduce autophagy when amino acids are restricted. A census of proteins degraded and fueled by autophagy shows how autophagy shapes CD8 T cell proteomes. In cytotoxic T cells, dominant autophagy substrates include cytolytic effector molecules, and amino acid and glucose transporters. In naive T cells, mitophagy dominates and selective mitochondrial pruning supports the expression of molecules that coordinate T cell migration and survival. Autophagy thus differentially prunes naive and effector T cell proteomes and is dynamically repressed by antigen receptors and inflammatory cytokines to shape T cell differentiation. Sinclair et al. combine quantitative proteomics and an autophagy flux reporter to map autophagy substrates and triggers during CD8 T cell differentiation. Proteins degraded and fueled by autophagy in naïve and effector T cells are identified and it is revealed that the regulation of amino acid transport allows antigen receptors and inflammatory cytokines to repress autophagy flux to shape T cell differentiation.

抗原和细胞因子对自噬的抑制影响了CD8 T细胞的线粒体、迁移和效应机制
自噬影响CD8 T细胞的命运;然而,这一进程的时机、触发因素和目标都没有明确定义。在此,我们发现初始CD8 T细胞具有高自噬通量,并且我们确定了一个自噬检查点,通过抗原受体接合和炎症细胞因子通过调节氨基酸转运蛋白表达和细胞内氨基酸递送来急性抑制自噬。具有高水平氨基酸转运体的活化T细胞在氨基酸充满的条件下具有低自噬通量,但当氨基酸受到限制时,可迅速恢复自噬。对自噬降解和促进的蛋白质的普查显示了自噬如何塑造CD8 T细胞蛋白质组。在细胞毒性T细胞中,主要的自噬底物包括细胞溶解效应分子、氨基酸和葡萄糖转运蛋白。在初始T细胞中,线粒体自噬占主导地位,选择性线粒体修剪支持协调T细胞迁移和存活的分子的表达。因此,自噬会对初始和效应T细胞蛋白质组进行不同的修剪,并受到抗原受体和炎症细胞因子的动态抑制,从而形成T细胞分化。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


