Probiotic Antimicrobial Evaluation Via Real-Time Profiling of Bacterial Cell Proliferation Using Stochastic Kinetics
IF 8.2
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
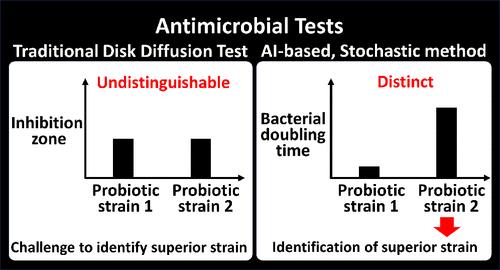
Probiotic metabolites are gaining attention as potential antibiotic candidates against antibiotic-resistant bacteria. The disk diffusion test, by measuring bacterial aggregate responses, faces challenges in accurately evaluating antimicrobial efficacy when these responses to different probiotic strains are indistinguishable at a macroscopic level. Here, this study presents an analytical method for accurately evaluating antimicrobial activity by analyzing bacterial cell proliferation suppression at a microscopic level. This assay can be used in a coculture system, designed to continuously expose pathogenic bacteria growing on the bottom surface of the culture plate to probiotic metabolites, selectively released from porous capsules positioned above. Bacterial proliferation is optically monitored in real-time and tracked via a computer vision algorithm. Specifically, bacterial proliferation is quantified as their doubling time, calculated using a proposed stochastic kinetic model. This method identifies the most potent antimicrobial strains by determining which probiotic candidates most effectively extend the bacterial doubling time. In comparative experiments using the same strains, this proposed method demonstrated clear distinctions in the antimicrobial efficacy of each strain, unlike the disk diffusion test. Therefore, this approach provides a reliable solution for identifying superior probiotic strains, with potential for widespread use in discovering new antimicrobial agents.
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Sensors
Chemical Engineering-Bioengineering
CiteScore
14.50
自引率
3.40%
发文量
372
期刊介绍:
ACS Sensors is a peer-reviewed research journal that focuses on the dissemination of new and original knowledge in the field of sensor science, particularly those that selectively sense chemical or biological species or processes. The journal covers a broad range of topics, including but not limited to biosensors, chemical sensors, gas sensors, intracellular sensors, single molecule sensors, cell chips, and microfluidic devices. It aims to publish articles that address conceptual advances in sensing technology applicable to various types of analytes or application papers that report on the use of existing sensing concepts in new ways or for new analytes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


