Using transcriptomics data and Adverse Outcome Pathway networks to explore endocrine disrupting properties of Cadmium and PCB-126
IF 10.3
1区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
Abstract
Omics-technologies such as transcriptomics offer valuable insights into toxicity mechanisms. However, integrating this type of data into regulatory frameworks remains challenging due to uncertainties regarding toxicological relevance and links to adverse outcomes. Furthermore, current assessments of endocrine disruptors (EDs) relevant to human health require substantial amounts of data, and primarily rely on standardized animal studies. Identifying EDs is a high priority in the EU, but so are efforts to replace and reduce animal testing. Alternative methods to investigate EDs are needed, and so are health risk assessment methods that support uptake of novel mechanistic information.
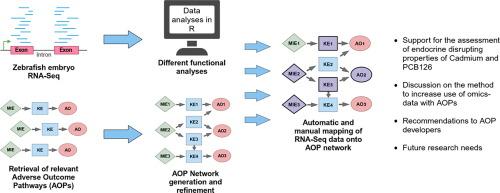
This study aims to utilize Adverse Outcome Pathways (AOPs) to integrate transcriptomics data for identifying EDs, by establishing a link between molecular data and adverse outcomes. Cadmium (Cd) and 3,3′,4,4′,5-pentachlorobiphenyl (PCB126) were used as model compounds due to their observed effects on the endocrine system. An AOP network for the estrogen, androgen, thyroid, and steroidogenesis (EATS)-modalities was constructed. RNA sequencing (RNA-Seq) was conducted on zebrafish (Danio rerio) embryos exposed to Cd or PCB126 for 4 days. RNA-Seq data were then linked to the AOP network via Gene Ontology (GO) terms. Enrichment Maps in Cytoscape and the QIAGEN Ingenuity Pathway Analysis (IPA) software were also used to identify potential ED properties and to support the assessment.
Potentially EATS-related GO Biological Process (BP) terms were identified for both compounds. A lack of accurate standardized terms in KEs of the AOP network hindered a data-driven mapping approach. Instead, manual mapping of GO BP terms onto the AOP network revealed more connections, underscoring the need for harmonizing AOP development for regulatory use. Both the Enrichment Maps and the IPA results further supported potentially EATS-related effects of both compounds. While AOP networks show promise in integrating RNA-Seq data, several challenges remain.

求助全文
约1分钟内获得全文
求助全文
来源期刊

Environment International
环境科学-环境科学
CiteScore
21.90
自引率
3.40%
发文量
734
审稿时长
2.8 months
期刊介绍:
Environmental Health publishes manuscripts focusing on critical aspects of environmental and occupational medicine, including studies in toxicology and epidemiology, to illuminate the human health implications of exposure to environmental hazards. The journal adopts an open-access model and practices open peer review.
It caters to scientists and practitioners across all environmental science domains, directly or indirectly impacting human health and well-being. With a commitment to enhancing the prevention of environmentally-related health risks, Environmental Health serves as a public health journal for the community and scientists engaged in matters of public health significance concerning the environment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


