Systematic reconstruction of molecular pathway signatures using scalable single-cell perturbation screens
IF 19.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
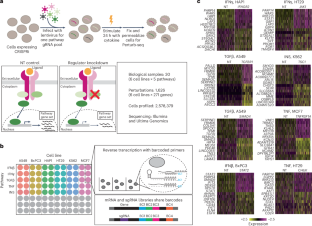
Recent advancements in functional genomics have provided an unprecedented ability to measure diverse molecular modalities, but predicting causal regulatory relationships from observational data remains challenging. Here, we leverage pooled genetic screens and single-cell sequencing (Perturb-seq) to systematically identify the targets of signalling regulators in diverse biological contexts. We demonstrate how Perturb-seq is compatible with recent and commercially available advances in combinatorial indexing and next-generation sequencing, and perform more than 1,500 perturbations split across six cell lines and five biological signalling contexts. We introduce an improved computational framework (Mixscale) to address cellular variation in perturbation efficiency, alongside optimized statistical methods to learn differentially expressed gene lists and conserved molecular signatures. Finally, we demonstrate how our Perturb-seq derived gene lists can be used to precisely infer changes in signalling pathway activation for in vivo and in situ samples. Our work enhances our understanding of signalling regulators and their targets, and lays a computational framework towards the data-driven inference of an ‘atlas’ of perturbation signatures. In two independent studies, Song et al. and Jiang, Dalgarno et al. present computational frameworks, perturbation-response score and Mixscale, respectively, to determine individual cellular variation in response to perturbation.

利用可扩展的单细胞摄动屏幕系统重建分子通路特征
功能基因组学的最新进展提供了前所未有的测量不同分子模式的能力,但从观察数据预测因果调节关系仍然具有挑战性。在这里,我们利用汇集的遗传筛选和单细胞测序(Perturb-seq)来系统地识别不同生物学背景下信号调节因子的靶标。我们展示了Perturb-seq如何与组合索引和下一代测序的最新和商业上可用的进展兼容,并在6个细胞系和5个生物信号环境中进行了超过1500次的扰动。我们引入了一个改进的计算框架(Mixscale)来解决细胞在扰动效率方面的变化,同时优化了统计方法来学习差异表达的基因列表和保守的分子特征。最后,我们展示了如何使用我们的Perturb-seq衍生基因列表来精确推断体内和原位样品信号通路激活的变化。我们的工作增强了我们对信号调节器及其目标的理解,并为扰动特征的“图谱”的数据驱动推断奠定了计算框架。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


