Decoding heterogeneous single-cell perturbation responses
IF 19.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
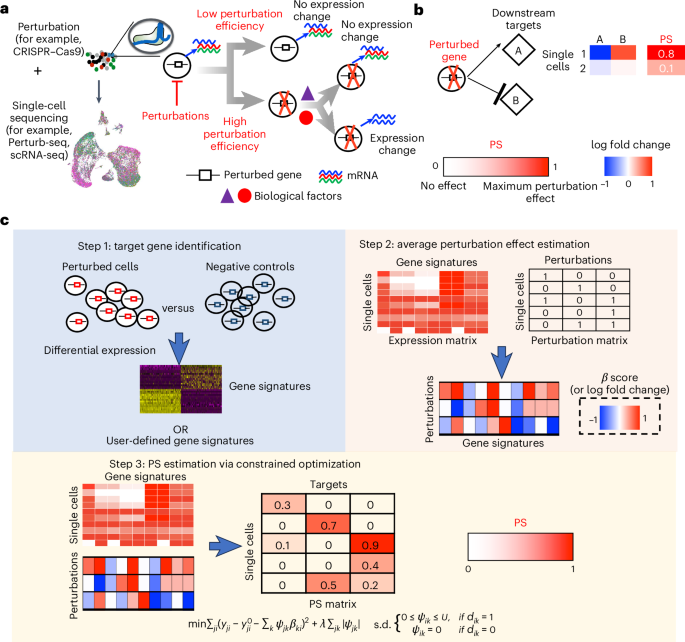
Understanding how cells respond differently to perturbation is crucial in cell biology, but existing methods often fail to accurately quantify and interpret heterogeneous single-cell responses. Here we introduce the perturbation-response score (PS), a method to quantify diverse perturbation responses at a single-cell level. Applied to single-cell perturbation datasets such as Perturb-seq, PS outperforms existing methods in quantifying partial gene perturbations. PS further enables single-cell dosage analysis without needing to titrate perturbations, and identifies ‘buffered’ and ‘sensitive’ response patterns of essential genes, depending on whether their moderate perturbations lead to strong downstream effects. PS reveals differential cellular responses on perturbing key genes in contexts such as T cell stimulation, latent HIV-1 expression and pancreatic differentiation. Notably, we identified a previously unknown role for the coiled-coil domain containing 6 (CCDC6) in regulating liver and pancreatic cell fate decisions. PS provides a powerful method for dose-to-function analysis, offering deeper insights from single-cell perturbation data. In two independent studies, Song et al. and Jiang, Dalgarno et al. present computational frameworks, perturbation-response score and Mixscale, respectively, to determine individual cellular variation in response to perturbation.

解码异质单细胞扰动响应
了解细胞对扰动的不同反应在细胞生物学中是至关重要的,但现有的方法往往不能准确地量化和解释异质单细胞反应。在这里,我们介绍了扰动响应评分(PS),一种在单细胞水平上量化不同扰动响应的方法。应用于单细胞扰动数据集,如Perturb-seq, PS在量化部分基因扰动方面优于现有方法。PS进一步使单细胞剂量分析不需要滴定扰动,并确定“缓冲”和“敏感”的基本基因的反应模式,这取决于它们的适度扰动是否导致强烈的下游效应。PS揭示了在T细胞刺激、潜伏HIV-1表达和胰腺分化等情况下,细胞对干扰关键基因的差异反应。值得注意的是,我们发现了含有6的螺旋结构域(CCDC6)在调节肝脏和胰腺细胞命运决定中的先前未知的作用。PS提供了一种强大的剂量-功能分析方法,从单细胞扰动数据中提供更深入的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


