Pre-existing stem cell heterogeneity dictates clonal responses to the acquisition of leukemic driver mutations
IF 19.8
1区 医学
Q1 CELL & TISSUE ENGINEERING
引用次数: 0
Abstract
Cancer cells display wide phenotypic variation even across patients with the same mutations. Differences in the cell of origin provide a potential explanation, but traditional assays lack the resolution to distinguish clonally heterogeneous subsets of stem and progenitor cells. To address this challenge, we developed simultaneous tracking of recombinase activation and clonal kinetics (STRACK), a method to trace clonal dynamics and gene expression before and after the acquisition of cancer mutations. Using mouse models, we studied two leukemic mutations, Dnmt3a-R878H and Npm1c, and found that their effect was highly variable across different stem cell states. Specifically, a subset of differentiation-primed stem cells, which normally becomes outcompeted with time, expands with both mutations. Intriguingly, Npm1c mutations reversed the intrinsic bias of the clone of origin, with differentiation-primed stem cells giving rise to more primitive malignant states. Thus, we highlight the relevance of single-cell lineage tracing to unravel early events in cancer evolution and posit that different cellular histories carry distinct cancer phenotypic potential.
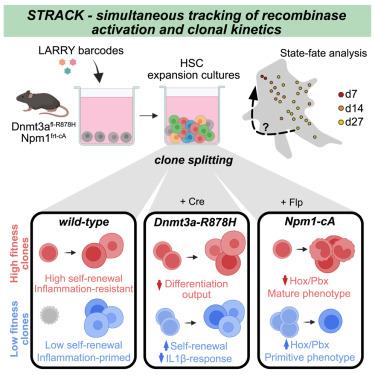
预先存在的干细胞异质性决定了对白血病驱动突变获得的克隆反应
即使在具有相同突变的患者之间,癌细胞也显示出广泛的表型差异。细胞来源的差异提供了一种可能的解释,但传统的检测方法缺乏区分干细胞和祖细胞的克隆异质亚群的分辨率。为了解决这一挑战,我们开发了同步跟踪重组酶激活和克隆动力学(STRACK),这是一种追踪癌症突变获得前后克隆动力学和基因表达的方法。利用小鼠模型,我们研究了两种白血病突变,Dnmt3a-R878H和Npm1c,发现它们的影响在不同的干细胞状态下变化很大。具体来说,分化引发的干细胞的一个子集,通常会随着时间的推移而失去竞争优势,随着这两种突变而扩大。有趣的是,Npm1c突变逆转了起源克隆的固有偏见,分化引发的干细胞产生更原始的恶性状态。因此,我们强调单细胞谱系追踪的相关性,以揭示癌症进化的早期事件,并假设不同的细胞历史携带不同的癌症表型潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell stem cell
生物-细胞生物学
CiteScore
37.10
自引率
2.50%
发文量
151
审稿时长
42 days
期刊介绍:
Cell Stem Cell is a comprehensive journal covering the entire spectrum of stem cell biology. It encompasses various topics, including embryonic stem cells, pluripotency, germline stem cells, tissue-specific stem cells, differentiation, epigenetics, genomics, cancer stem cells, stem cell niches, disease models, nuclear transfer technology, bioengineering, drug discovery, in vivo imaging, therapeutic applications, regenerative medicine, clinical insights, research policies, ethical considerations, and technical innovations. The journal welcomes studies from any model system providing insights into stem cell biology, with a focus on human stem cells. It publishes research reports of significant importance, along with review and analysis articles covering diverse aspects of stem cell research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


