Thinking on the Use of Artificial Intelligence in Drug Discovery
IF 6.8
1区 医学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
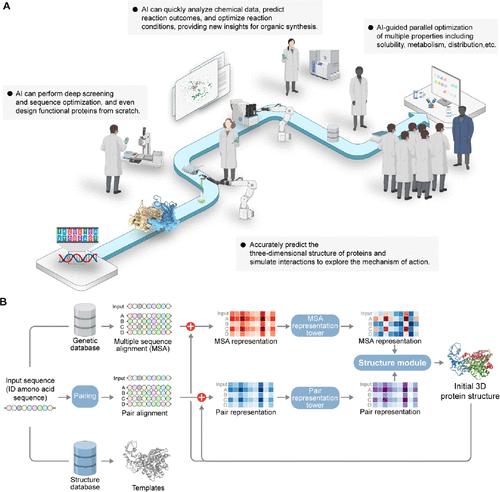
Advances in machine learning algorithms and big data processing capabilities have propelled artificial intelligence (AI) to the forefront, with its applications in drug discovery rapidly increasing. A report from McKinsey & Company indicates that the use of AI can help pharmaceutical companies save 30 to 50% in drug development costs and increase the pipeline speed by over 20%. (1) AI technology is becoming increasingly common from drug discovery to clinical trials. First, AI can quickly screen potential active compounds using machine learning and deep learning algorithms, guiding hit-to-lead optimization to improve therapeutic efficacy and reduce toxicity, significantly enhancing the efficiency of drug discovery. Second, AI-driven predictive models can enhance the success rate of experiments, reduce research and development (R&D) costs, and even change a drug’s behavior in the body, including its pharmacokinetics (PK) and pharmacodynamics (PD), to optimize dosage and dosing regimens. By integrating expertise from various disciplines, it enables the derivation of novel conclusions, producing forward-looking results in traditionally empirical areas. Furthermore, AI has driven the development of personalized medicine, bringing significant transformations and opportunities for the future of healthcare. Drug discovery and development is the most advanced area of AI, with numerous breakthroughs already achieved (Figure 1A). For instance, Alex Zhavoronkov’s team developed a deep generative model, generative tensorial reinforcement learning (GENTRL) for de novo small molecule design. (2) The model combines reinforcement learning, variational inference, and tensor decompositions for rapid generation and optimization of potentially pharmacologically active compounds. GENTRL uses three layers of self-organizing mappings (SOMs) to generate innovative DDR1 inhibitors, and these small molecules then undergo multiple screenings and optimizations, resulting in six candidate compounds in just 21 days, with four showing potent biological activity. Even more surprisingly, ISM001-055AI, a first-in-class small molecule inhibitor developed by generative AI technology, shows positive results in a phase IIa clinical trial for treating idiopathic pulmonary fibrosis (IPF). (3) In September 2024, Insilico Medicine announced the results of a Phase IIa clinical trial of ISM001-055. The trial, which enrolled 71 IPF patients across 21 clinical research centers in China, demonstrated that ISM001-055 had a favorable safety profile at all dose levels and exhibited a dose-dependent trend of efficacy in forced vital capacity (FVC), an important measure of lung function in IPF patients. The improvement in lung function in just 12 weeks brings great hope to IPF patients. The success of ISM001-055 showcases the huge potential of AI in drug discovery and development, paving the way for larger-scale trials in the future. Fang’s team recently developed a robotic system for high-throughput chemical synthesis, online characterization, and large-scale photocatalytic reaction condition screening (10,000 reaction conditions per day). The system is based on liquid-core waveguide, microfluidic liquid-handling, and artificial intelligence techniques to significantly reduce the time and effort required for complex organic synthesis. (4) They have also developed an AI-assisted method for predicting absorbance, analyzing factors that influence convective and molecular diffusion effects. When a candidate compound is selected at the chemistry, manufacturing, and control (CMC) stage, crystal structure prediction (CSP) and structure-based stability evaluation can be used with advanced experimental screening techniques to minimize the risk of polymorphic problems and accelerate decision-making for formulation studies. (5,6) During the development of Paxlovid, Pfizer’s oral Covid-19 treatment, Pfizer and XtalPi elucidated the subtle differences in structure, dynamics, and stability of two enantiotropically related anhydrous polymorphs of the novel antiviral medicine nirmatrelvir by employing a series of highly orthogonal experimental and computational approaches. (7) This work ultimately determined Form 1 for formulation development as it is the stable form above the transition temperature of 17 °C, demonstrating the unprecedented speed in bringing Paxlovid to patients in record time amidst the pandemic. Figure 1. (A) AI-driven drug discovery process, including de novo design, compound screening, interaction mulations, organic synthesis, and optimization of pharmacokinetic properties. (B) The AlphaFold model architecture predicts the initial protein structure, demonstrating near-experimental accuracy in protein structure prediction through recycling (PDB code: 6Y4F). Some elements were created with BioRender (https://biorender.com). Antibody drugs, along with small molecule drugs, are widely used in treating cancer, autoimmune diseases, and infectious diseases because of their high specificity for targets. (8) AI can be used for structure prediction, design optimization, humanization of antibody drugs, and predicting the optimal drug-to-antibody ratio (DAR) for antibody-drug conjugates (ADCs). This helps shorten development timelines and increase success rates. AI’s superior generalization ability and high degree of automation make it highly effective in vaccine development, which usually takes over 10 years. For example, AI has played a crucial role in optimizing mRNA sequences. (9,10) The LinearDesign deep learning algorithm has greatly enhanced the stability, protein expression, and immunogenicity of mRNA vaccines by optimizing mRNA sequence structural stability and codon usage. The algorithm successfully solved the large-scale search problem in traditional mRNA design, finding the optimal mRNA sequence in less than 11 min. It showcases the potential of AI and computational linguistics technologies in mRNA design and was licensed to Moderna, a leading global RNA technology company in 2022 to enhance the development of mRNA vaccines and therapeutics. (10) AlphaFold is a recent breakthrough in protein structure prediction that uses a neural network-based machine learning method. It combines evolutionary data and geometric properties to accurately predict three-dimensional structures (Figure 1B). (9) After entering the amino acid sequence of a protein, Evoformer generates a multiple sequence alignment (MSA) and templates from relevant evolutionary databases to capture the relationships and covariation between sequences. The predicted structure is then fed back into Evoformer and the structure module multiple times for iterative optimization. Each recursive step, known as “Recycling,” refines the structure further to enhance accuracy. Significant progress has been made in predicting protein structures, but this is only the beginning of exploration in life sciences. AI is currently mainly used at the molecular level, but future research will integrate diverse high-quality data and advance to higher levels to discover new patterns and principles in biological systems. For example, the Peking University International Cancer Institute has partnered with XtalPi to investigate disease mechanisms and drug actions through multimodal data integration. (11) The Shu group at Peking University utilized high-throughput CRISPR technology for large-scale gene editing in cells, whereas XtalPi’s independently developed cell research platform, X-Map, which allows for collection of large-scale, high-content imaging and transcriptome data following cell perturbation. The X-Profiler algorithm developed by XtalPi is proficient at extracting relevant information for specific downstream tasks. It effectively addresses issues like out-of-focus blurring in high-content imaging due to variations in well plate edge height, reduces data noise, and enhances the signal-to-noise ratio (SNR). Furthermore, the X-Profiler can adaptively adjust data quality control strategies based on task requirements, significantly boosting model performance. By integrating experimental methods with AI algorithms, this approach enables precise observations at the cellular level using real-world multidimensional data, establishing correlations between physiological changes and gene or drug regulation. This method offers higher throughput and lower costs compared to animal models, allowing for quicker generation of high-quality data for specific research systems and improving the efficiency and success rate of drug discovery and development. Although AI is well established in many aspects of drug discovery and development, the outdated traditional education system is no longer suitable for the rapid development of AI and urgently needs reform and optimization. To address the educational challenges posed by the rapid development of AI, it is essential to have a clear understanding of its specific applications. This will help us pinpoint which areas of teaching content require additional support. The traditional teaching methods have issues like outdated curricula, lack of practical experience, and limited interdisciplinary integration, focusing too much on foundational subjects and neglecting crucial skills in data science and AI. The lack of interdisciplinary integration and practical experience makes it difficult to meet the demands of the AI era. Given AI’s increasing importance, teaching approaches should be updated. First and foremost, teachers should be trained with AI technologies to ensure they are prepared to teach AI courses. Second, interdisciplinary integration in education should be promoted to inspire insights from different disciplines. Computer science and data science should be integrated into the pharmaceutical curriculum to break down disciplinary silos. For example, the basic theory component of the drug molecular design course should include fundamental concepts of AI, machine learning, and deep learning, along with their applications in the pharmaceutical industry. This will cover skills such as data preprocessing, model training and evaluation, and feature engineering. The core course section should explore AI applications in drug target screening, molecular docking, virtual screening, molecular structure analysis, and AI modeling comprehensively. Cutting-edge AI technologies in drug discovery, such as reinforcement learning and Generative Adversarial Networks (GANs), should be introduced. GANs are a specialized type of deep learning with two neural networks─one for image generation and one for discrimination. They can efficiently create diverse, high-quality molecules, showing unique advantages and potential by learning existing data distribution to explore new chemical spaces. Third, new courses should be added and professors from various fields should collaborate to edit textbooks that combine pharmaceutical science with AI for the updated curriculum. International academic conferences and online courses are also valuable educational resources for students to learn AI. It is important to expand AI-related courses, including big data analysis, natural language processing, bioinformatics, etc., to enhance relevant skills. However, increasing reliance on AI learning unilaterally will only add more pressure on students, thus learning new courses should be personalized to avoid overwhelming students with the broad scope of AI content. Reform should focus on innovation, professionalism, and personalization to better address individual learning needs. Students should choose courses based on their professional needs, focusing on relevant skills after gaining foundational knowledge in the early stages of education. Use AI technology in real-world drug development scenarios to teach students how to solve practical problems. AI-driven practical training and hands-on experience must be included. For example, creating a virtual lab platform that integrates AI models and data analysis tools for simulating real-world scenarios like drug screening, molecular docking, and drug safety prediction. This platform should also have online collaboration features to allow students to work together in real time, mimicking a professional work environment that promotes interdisciplinary teamwork. Additionally, enhancing partnerships with pharmaceutical and AI technology companies to develop a platform that connects industry and academia is crucial. Project content should be tailored to meet the actual needs of industry partners to offer students practical solutions relevant to the field. By gaining a strong grasp of AI concepts, students will be better prepared for future roles in drug development. To be expected, AI technology has and will profoundly revolutionize the field of drug discovery. Educational policymakers and academic institutions globally should update their training programs to incorporate advanced AI technologies. Students need to learn latest AI techniques like deep learning and natural language processing to be better prepared for future challenges in drug development. Students without AI skills can be readily replaced by those who embrace AI advancement. However, we have to keep in mind that AI may not be a panacea due to its limitation in the quality of the data. High-quality, comprehensive experimental data sets are essential for generating reliable results, yet not all entities with an AI platform possess such data. The impact of AI and machine learning in drug discovery is directly dependent on the quality and depth of the training data used to develop the AI models. Regardless of how advanced the algorithms are, if the training data is biased or of poor quality, the AI predictions will be inaccurate, thereby limiting their potential to drive new drug discovery. And algorithmic biases can lead to inaccurate therapeutic effects of the model on certain populations, which is especially critical in the safety of patients’ medication. Additionally, it is not only important to pay attention to the quality of the data, but also to ensure the protection of sensitive patient data. We need to establish strict data protection measures to ensure the security of the data during acquisition, storage, and processing, thereby safeguarding patient data privacy. Y.W. and Z.H. contributed equally to this work. This work was provided by the National Natural Science Foundation of China (Nos. 82473782, 22277110, and 82473761), Sichuan Science and Technology Program (No. 2024ZYD0130), Natural Science Foundation of Henan Province (No. 242301420005), Key Research Project for Basic Research in Henan Province Universities (No. 25ZX001). The authors acknowledge Professor Craig W. Lindsley (Editor-in-Chief of Journal of Medicinal Chemistry; Executive Director, Warren Center for Neuroscience Drug Discovery, Vanderbilt University) and Dr. Shuhao Wen (Chairman of XtalPi) for their insightful discussions. This article references 11 other publications. This article has not yet been cited by other publications.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Medicinal Chemistry
医学-医药化学
CiteScore
4.00
自引率
11.00%
发文量
804
审稿时长
1.9 months
期刊介绍:
The Journal of Medicinal Chemistry is a prestigious biweekly peer-reviewed publication that focuses on the multifaceted field of medicinal chemistry. Since its inception in 1959 as the Journal of Medicinal and Pharmaceutical Chemistry, it has evolved to become a cornerstone in the dissemination of research findings related to the design, synthesis, and development of therapeutic agents.
The Journal of Medicinal Chemistry is recognized for its significant impact in the scientific community, as evidenced by its 2022 impact factor of 7.3. This metric reflects the journal's influence and the importance of its content in shaping the future of drug discovery and development. The journal serves as a vital resource for chemists, pharmacologists, and other researchers interested in the molecular mechanisms of drug action and the optimization of therapeutic compounds.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


