Fast screening of COVID-19 inpatient samples by integrating machine learning and label-free SERS methods
IF 5.7
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
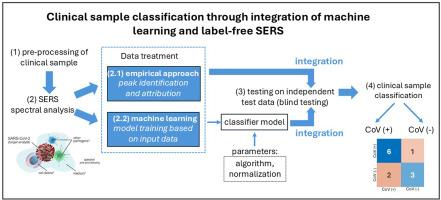
Advances in bio-analyte detection demonstrate the need for innovation to overcome the limitations of traditional methods. Emerging viruses evolve into variants, driving the need for fast screening to minimize the time required for positive detection and establish standardized detection. In this study, a SERS-active substrate with Au NPs on a regularly arranged ZrO2 nanoporous structure was utilized to obtain the SERS spectrum of inpatient samples from COVID-19 patients. Two analytical approaches were applied to classify clinical samples - empirical method to identify peak assignments corresponding to the target SARS-CoV-2 BA.2 variant, and machine learning (ML) method to build classifier models.
Results
Comparison of spectral profiles of pure BA.2 variant and inpatient samples showed significant differences in the occurrence of SERS peaks, requiring the selection of regions of interest for further analysis through the empirical method. SERS spectra are classified into CoV (+) and CoV (−) using both empirical and machine learning methods, each demonstrating a sensitivity of 85.7 % and a specificity of 60 %. The former method relies on peak assignment, which is performed manually relying on established and results in a longer turnaround time. In contrast, the latter method is based on the mathematical correlations between variables across the entire spectrum. The machine must continuously learn from larger datasets, and the response time for interpretation is short. Nonetheless, both methods demonstrated their suitability in classifying clinical samples.
Significance
This study demonstrated that a more comprehensive discussion can be formed with the integration of machine learning classification with biochemical profiling with the empirical analysis approach. Further improvement is expected by combining these two methods by utilizing only the regions of interest instead of the entire spectrum as input for machine learning, as a feature extraction technique.

求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


