De novo design of transmembrane fluorescence-activating proteins
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
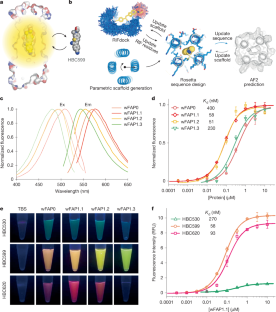
The recognition of ligands by transmembrane proteins is essential for the exchange of materials, energy and information across biological membranes. Progress has been made in the de novo design of transmembrane proteins1–6, as well as in designing water-soluble proteins to bind small molecules7–12, but de novo design of transmembrane proteins that tightly and specifically bind to small molecules remains an outstanding challenge13. Here we present the accurate design of ligand-binding transmembrane proteins by integrating deep learning and energy-based methods. We designed pre-organized ligand-binding pockets in high-quality four-helix backbones for a fluorogenic ligand, and generated a transmembrane span using gradient-guided hallucination. The designer transmembrane proteins specifically activated fluorescence of the target fluorophore with mid-nanomolar affinity, exhibiting higher brightness and quantum yield compared to those of enhanced green fluorescent protein. These proteins were highly active in the membrane fraction of live bacterial and eukaryotic cells following expression. The crystal and cryogenic electron microscopy structures of the designer protein–ligand complexes were very close to the structures of the design models. We showed that the interactions between ligands and transmembrane proteins within the membrane can be accurately designed. Our work paves the way for the creation of new functional transmembrane proteins, with a wide range of applications including imaging, ligand sensing and membrane transport. A study describes the design of de novo ligand-binding transmembrane proteins, demonstrating their specific binding and activation of fluorogenic ligands.

跨膜荧光激活蛋白的从头设计
跨膜蛋白对配体的识别对于跨生物膜的物质、能量和信息交换至关重要。跨膜蛋白1、2、3、4、5、6的从头设计,以及设计与小分子结合的水溶性蛋白7、8、9、10、11、12,都取得了进展,但从头设计与小分子紧密特异性结合的跨膜蛋白仍然是一个突出的挑战13。在这里,我们通过整合深度学习和基于能量的方法,提出了配体结合跨膜蛋白的精确设计。我们在高质量的四螺旋骨架中设计了预先组织的配体结合袋,用于荧光配体,并使用梯度引导幻觉产生跨膜跨度。设计的跨膜蛋白特异性激活了目标荧光团的荧光,具有中等纳摩尔亲和力,与增强的绿色荧光蛋白相比,具有更高的亮度和量子产率。表达后,这些蛋白在活细菌和真核细胞的膜组分中具有高度活性。设计蛋白-配体复合物的晶体和低温电镜结构与设计模型的结构非常接近。我们发现配体与膜内跨膜蛋白之间的相互作用可以被精确地设计。我们的工作为创造新的功能性跨膜蛋白铺平了道路,具有广泛的应用,包括成像,配体传感和膜运输。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


