Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
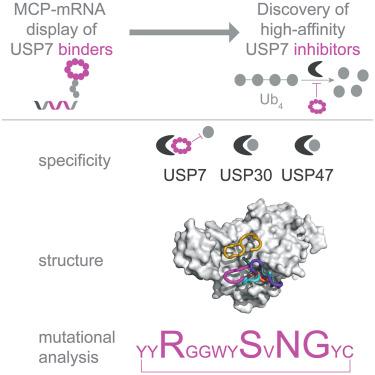
The ubiquitin-specific protease (USP) family of deubiquitinases (DUBs) are regulators of Ub signaling that share a common catalytic-domain fold. The dynamic nature of this domain is important for controlling the function of USPs, with inter- and intramolecular interactions often influencing the structure and enzymatic activity of these DUBs. This conformational flexibility, in combination with the high sequence conservation of the USP active site, has made it challenging to readily identify potent and selective inhibitors for individual USPs. Here, we demonstrate how a naive, macrocycle-mRNA display selection rapidly yielded high-affinity binders to USP7 that specifically inhibit the DUB with nanomolar half-maximal inhibitory concentration (IC50) values. Structural analysis of the macrocycles bound to USP7 revealed a variety of binding modes and identified inhibition hotspots on the enzyme that mirror those used by small-molecule inhibitors. Together, these data suggest that initial macrocyclic hits could serve as pivotal tools in developing USP-specific inhibitors and probing USP biology.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


