Biases in machine-learning models of human single-cell data
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
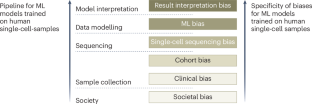
Recent machine-learning (ML)-based advances in single-cell data science have enabled the stratification of human tissue donors at single-cell resolution, promising to provide valuable diagnostic and prognostic insights. However, such insights are susceptible to biases. Here we discuss various biases that emerge along the pipeline of ML-based single-cell analysis, ranging from societal biases affecting whose samples are collected, to clinical and cohort biases that influence the generalizability of single-cell datasets, biases stemming from single-cell sequencing, ML biases specific to (weakly supervised or unsupervised) ML models trained on human single-cell samples and biases during the interpretation of results from ML models. We end by providing methods for single-cell data scientists to assess and mitigate biases, and call for efforts to address the root causes of biases. This Perspective discusses the various biases that can emerge along the pipeline of machine learning-based single-cell analysis and presents methods to train models on human single-cell data in order to assess and mitigate these biases.

求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


