Innovative Multiplex qPCR Method for Rapid and Reliable Detection of Microcystin-Producing Genes during Harmful Algal Blooms: Insights from Utah Reservoirs
IF 11.4
1区 环境科学与生态学
Q1 ENGINEERING, ENVIRONMENTAL
引用次数: 0
Abstract
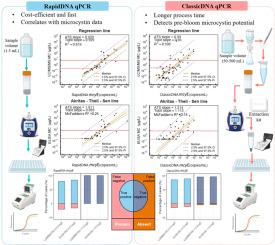
Cyanobacterial harmful algal blooms (cyanoHABs) have the potential to produce cyanotoxins, which pose significant health risks to both humans and animals. The gold standard methods for monitoring cyanoHABs involve enzyme-linked immunosorbent assay (ELISA), liquid chromatography combined with triple quadrupole mass spectrometry (LC-MS/MS) and manual cell counting under microscopy. However, these techniques, while effective, are costly and time-consuming, which may not be optimal for timely decision-making to safeguard public health. Quantitative polymerase chain reaction (qPCR) offers a complementary approach that serves as an indicator of the potential for toxin production. It provides accurate results with a rapid turnaround time and high throughput capacity, and greater affordability. To assess the reliability of qPCR in predicting toxin production and determining when toxin levels exceed recreational advisory thresholds, we conducted experiments utilizing two DNA extraction methods for qPCR testing: RapidDNA and ClassicDNA. Sampling was conducted across nine water bodies in Utah throughout the recreational season from June 1 to October 31, 2023. We targeted cyanotoxin-encoding genes mainly associated with microcystins, the dominant cyanotoxin reported for these water bodies, for qPCR analysis. Toxin levels were measured using both ELISA and LC-MS/MS with cyanobacteria cell counting conducted as a reference. Out of nine water bodies studied, cyanoHABs were detected in five (i.e., Utah Lake, and Deer Creek, Echo, Schofield, and Pineview Reservoirs). Analysis of the data revealed a significant linear relationship between both the qPCR results of mcyE (associated with microcystin production) obtained from RapidDNA and ClassicDNA methods, and the levels of microcystins measured by ELISA and LC-MS/MS. RapidDNA qPCR methods offer a potential warning tool for indicating toxin production during blooming events, though this method is not suitable for determining risk during the pre-blooming period. Conversely, ClassicDNA methods can be utilized during the pre-blooming period to prepare for potential blooms. These results provide insight into the genetic potential of blooms around the state to produce microcystins. Findings can be implemented in both Recreational Water Quality and Drinking Water programs nationally.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Water Research
环境科学-工程:环境
CiteScore
20.80
自引率
9.40%
发文量
1307
审稿时长
38 days
期刊介绍:
Water Research, along with its open access companion journal Water Research X, serves as a platform for publishing original research papers covering various aspects of the science and technology related to the anthropogenic water cycle, water quality, and its management worldwide. The audience targeted by the journal comprises biologists, chemical engineers, chemists, civil engineers, environmental engineers, limnologists, and microbiologists. The scope of the journal include:
•Treatment processes for water and wastewaters (municipal, agricultural, industrial, and on-site treatment), including resource recovery and residuals management;
•Urban hydrology including sewer systems, stormwater management, and green infrastructure;
•Drinking water treatment and distribution;
•Potable and non-potable water reuse;
•Sanitation, public health, and risk assessment;
•Anaerobic digestion, solid and hazardous waste management, including source characterization and the effects and control of leachates and gaseous emissions;
•Contaminants (chemical, microbial, anthropogenic particles such as nanoparticles or microplastics) and related water quality sensing, monitoring, fate, and assessment;
•Anthropogenic impacts on inland, tidal, coastal and urban waters, focusing on surface and ground waters, and point and non-point sources of pollution;
•Environmental restoration, linked to surface water, groundwater and groundwater remediation;
•Analysis of the interfaces between sediments and water, and between water and atmosphere, focusing specifically on anthropogenic impacts;
•Mathematical modelling, systems analysis, machine learning, and beneficial use of big data related to the anthropogenic water cycle;
•Socio-economic, policy, and regulations studies.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


