Review Article: Novel Enzyme Therapy Design for Gluten Peptide Digestion Through Exopeptidase Supplementation
Abstract
Background
Dietary peptides are increasingly linked to inflammatory gastrointestinal diseases, exemplified by coeliac disease. Coeliac disease is caused by an acquired immune response to proline- and glutamine-rich gluten peptides, which bottleneck proteolysis and provide substrates for immune recognition. Enzyme therapies aim to eliminate gluten immunogenic peptides as an adjunct to gluten-free diet.
Aims
To investigate overlooked aspects of enzyme development given difficulties in translating preclinical efficacy into clinical benefit.
Methods
We assessed mode-of-action, target organ and drug delivery in the context of digestive physiology and motility for gluten-digesting enzymes on the market or in development until 1 December 2024.
Results
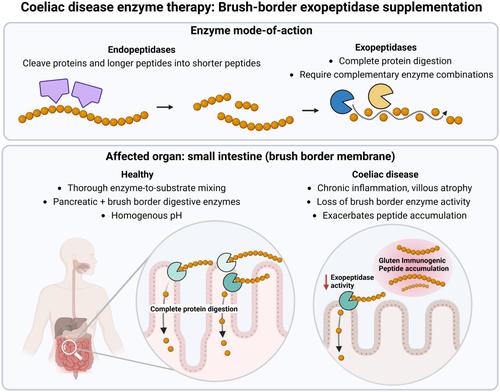
Most enzymes were gastric endopeptidases specific for proline or glutamine residues. Gastric enzymes may achieve poor enzyme–substrate exposure due to limited mixing and rapid emptying of water-soluble particles. Moreover, endopeptidases cleave proteins/peptides into shorter peptides but do not systematically cleave protein into absorbable fractions. Natural digestive physiology provides thorough mixing at the intestinal brush border, which produces exopeptidases necessary to fully digest proline-rich peptides. Despite reduced activity in patients with coeliac disease, exopeptidases remain underexplored as therapeutic agents. Given limited substrate scope and end-to-end digestion, exopeptidases are ineffective as single agents, requiring functional combinations. Furthermore, vulnerability to gastric acid requires stabilisation or formulation for rapid enteric release.
Conclusions
Enzymes should be stabilised throughout the gastrointestinal tract including the small intestine. Exopeptidases perform a critical function by systematically generating absorbable fractions, warranting future investigation as therapeutic agents. Sensitive and translational biomarkers are needed to better assess enzyme efficacy in real-meal conditions.


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


