Leveraging large language models for peptide antibiotic design.
IF 7.3
2区 综合性期刊
Q1 CHEMISTRY, MULTIDISCIPLINARY
Cell Reports Physical Science
Pub Date : 2025-01-15
Epub Date: 2024-12-31
DOI:10.1016/j.xcrp.2024.102359
引用次数: 0
Abstract
Large language models (LLMs) have significantly impacted various domains of our society, including recent applications in complex fields such as biology and chemistry. These models, built on sophisticated neural network architectures and trained on extensive datasets, are powerful tools for designing, optimizing, and generating molecules. This review explores the role of LLMs in discovering and designing antibiotics, focusing on peptide molecules. We highlight advancements in drug design and outline the challenges of applying LLMs in these areas.
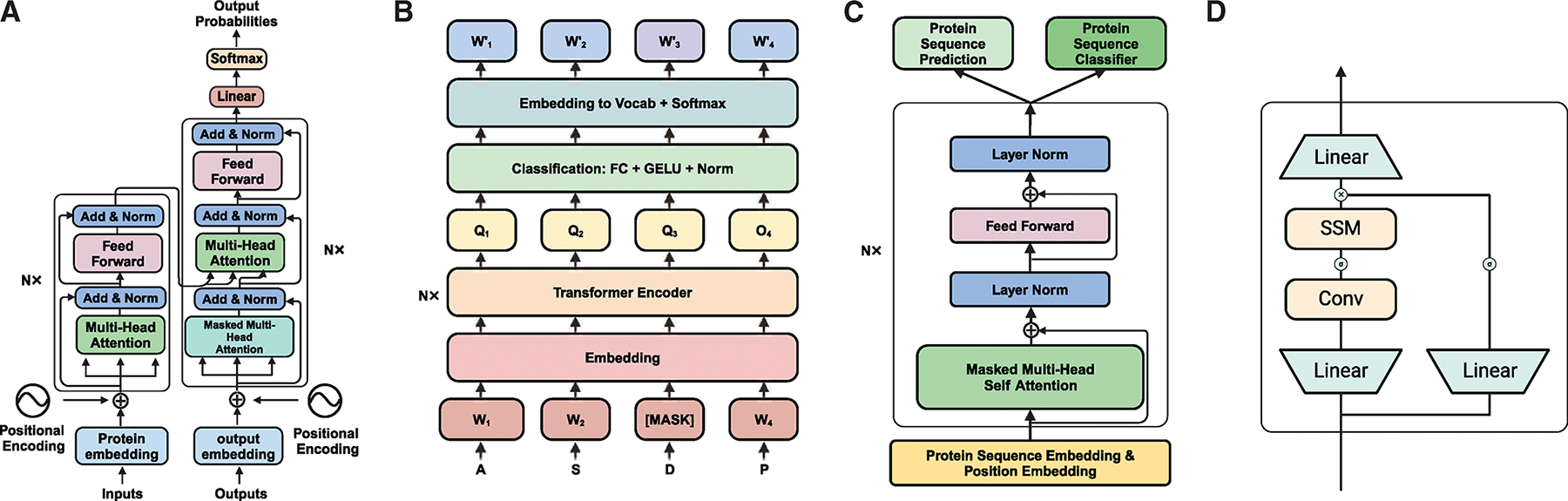
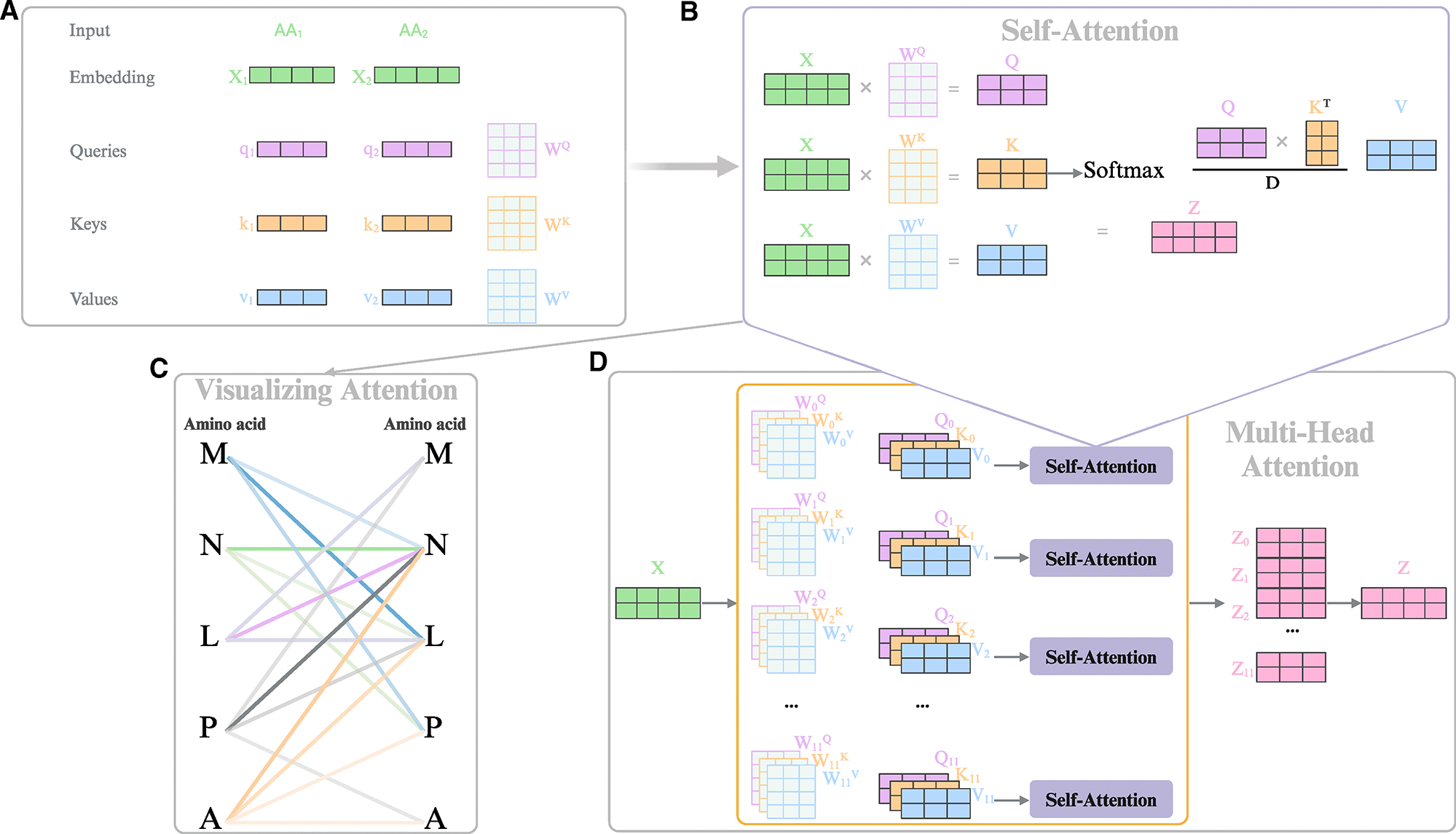
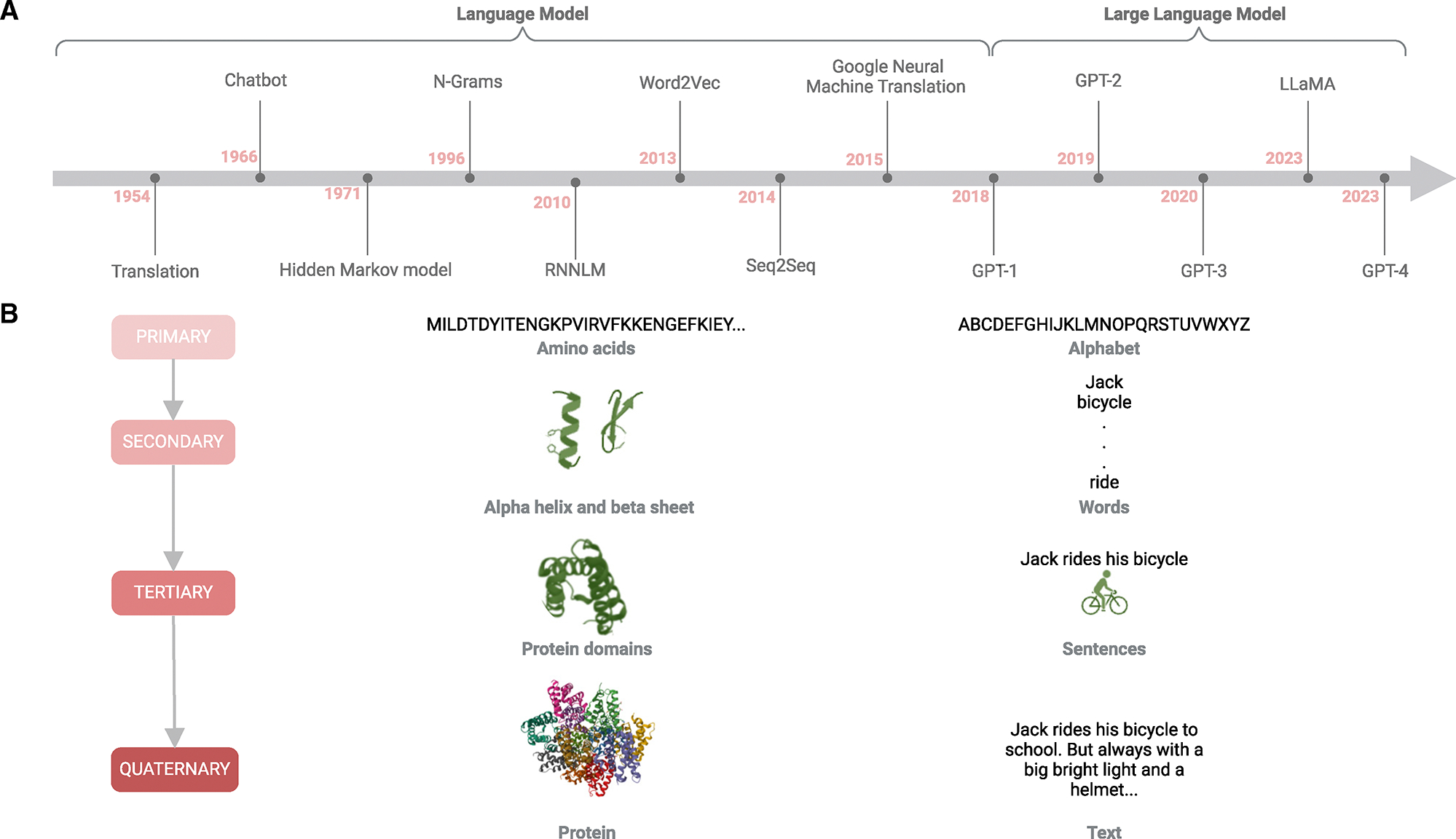
利用大型语言模型进行肽抗生素设计。
大型语言模型(llm)对我们社会的各个领域产生了重大影响,包括最近在生物和化学等复杂领域的应用。这些模型建立在复杂的神经网络架构上,并在广泛的数据集上进行训练,是设计、优化和生成分子的强大工具。本文综述了llm在发现和设计抗生素中的作用,重点是肽分子。我们强调了药物设计的进步,并概述了在这些领域应用法学硕士的挑战。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell Reports Physical Science
Energy-Energy (all)
CiteScore
11.40
自引率
2.20%
发文量
388
审稿时长
62 days
期刊介绍:
Cell Reports Physical Science, a premium open-access journal from Cell Press, features high-quality, cutting-edge research spanning the physical sciences. It serves as an open forum fostering collaboration among physical scientists while championing open science principles. Published works must signify significant advancements in fundamental insight or technological applications within fields such as chemistry, physics, materials science, energy science, engineering, and related interdisciplinary studies. In addition to longer articles, the journal considers impactful short-form reports and short reviews covering recent literature in emerging fields. Continually adapting to the evolving open science landscape, the journal reviews its policies to align with community consensus and best practices.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


