In situ lysosomal proteomics enabled by bioorthogonal photocatalytic proximity labelling
IF 44.6
1区 化学
Q1 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
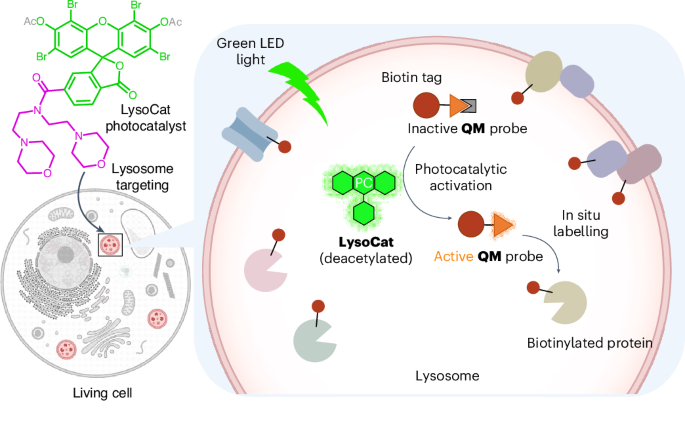
In situ deciphering of lysosome proteomes is crucial for understanding cellular processes and diseases, but is challenging due to its digestive, acidic environment that renders proximity labelling enzymes incompatible. Here we have developed a photocatalytic proximity labelling technique, CAT-Lyso, for in situ lysosomal proteomics. By employing a lysosome-targeting photocatalyst/thioquinone methide labelling probe pair, CAT-Lyso enables the generation of a reactive thioquinone methide intermediate via photoredox catalysis, facilitating efficient lysosomal proteome labelling in diverse cell lines, including hard-to-transfect macrophages (RAW264.7) and B lymphocytes (Raji). CAT-Lyso successfully identified cell type-specific lysosomal proteomic patterns and uncovered previously unrecognized lysosomal proteins, such as SCAMP3, NAGPA, GLG1 and MFSD14B. Furthermore, CAT-Lyso enabled quantitative profiling of lysosomal proteome dynamics under perturbations such as rapamycin-mediated mTOR inhibition, revealing pronounced ferritinophagy that evokes a coordinated labile iron-resisting program in cancer cells. With its in situ labelling, non-genetic operation, high specificity and photocontrollability, CAT-Lyso provides a powerful tool for investigating lysosome proteome dynamics in living systems. In situ profiling of lysosomal proteomes is impeded by the acidic and digestive environment of lysosomes. Now, a bioorthogonal photocatalytic system (CAT-Lyso) is developed that is compatible with these conditions, enabling the proximity labelling of lysosomal proteomes within living cells.

通过生物正交光催化接近标记实现原位溶酶体蛋白质组学
溶酶体蛋白质组的原位解码对于理解细胞过程和疾病至关重要,但由于其消化酸性环境使得接近标记酶不相容,因此具有挑战性。在这里,我们开发了一种光催化接近标记技术,CAT-Lyso,用于原位溶酶体蛋白质组学。CAT-Lyso采用一种靶向溶酶体的光催化剂/甲基硫醌标记探针对,能够通过光氧化还原催化生成活性的甲基硫醌中间体,促进溶酶体蛋白质组在多种细胞系中的高效标记,包括难以转染的巨噬细胞(RAW264.7)和B淋巴细胞(Raji)。CAT-Lyso成功鉴定了细胞类型特异性溶酶体蛋白质组学模式,并发现了以前未被识别的溶酶体蛋白,如SCAMP3, NAGPA, GLG1和MFSD14B。此外,CAT-Lyso能够在雷帕霉素介导的mTOR抑制等扰动下对溶酶体蛋白质组动力学进行定量分析,揭示了明显的铁蛋白自噬,在癌细胞中引发了协调的不稳定的抗铁程序。CAT-Lyso具有原位标记、非遗传操作、高特异性和光可控制性等优点,为研究生物系统中溶酶体蛋白质组动力学提供了强有力的工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Catalysis
Chemical Engineering-Bioengineering
CiteScore
52.10
自引率
1.10%
发文量
140
期刊介绍:
Nature Catalysis serves as a platform for researchers across chemistry and related fields, focusing on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, encompassing both fundamental and applied studies. With a particular emphasis on advancing sustainable industries and processes, the journal provides comprehensive coverage of catalysis research, appealing to scientists, engineers, and researchers in academia and industry.
Maintaining the high standards of the Nature brand, Nature Catalysis boasts a dedicated team of professional editors, rigorous peer-review processes, and swift publication times, ensuring editorial independence and quality. The journal publishes work spanning heterogeneous catalysis, homogeneous catalysis, and biocatalysis, covering areas such as catalytic synthesis, mechanisms, characterization, computational studies, nanoparticle catalysis, electrocatalysis, photocatalysis, environmental catalysis, asymmetric catalysis, and various forms of organocatalysis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


