A meta-analysis of the gut microbiome in inflammatory bowel disease patients identifies disease-associated small molecules
IF 20.6
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
Gut microbiome changes have been associated with several human diseases, but the molecular and functional details underlying these associations remain largely unknown. Here, we performed a meta-analysis of small molecule biosynthetic gene clusters (BGCs) in metagenomic samples of the gut microbiome from inflammatory bowel disease (IBD) patients and matched healthy subjects and identified two Clostridia-derived BGCs that are significantly associated with Crohn’s disease (CD), a main IBD type. Using synthetic biology, we discovered and solved the structures of six fatty acid amides as the products of the CD-enriched BGCs, which we subsequently detected in fecal samples from IBD patients. Finally, we show that the discovered molecules disrupt gut permeability and exacerbate disease in chemically or genetically susceptible mouse models of colitis. These findings suggest that microbiome-derived small molecules may play a role in the etiology of IBD and represent a generalizable approach for discovering molecular mediators of disease-relevant microbiome-host interactions.
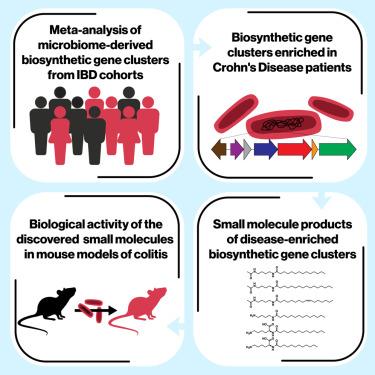
一项对炎症性肠病患者肠道微生物组的荟萃分析确定了与疾病相关的小分子
肠道微生物组的变化与几种人类疾病有关,但这些关联背后的分子和功能细节在很大程度上仍然未知。在这里,我们对炎症性肠病(IBD)患者和匹配的健康受试者的肠道微生物组宏基因组样本中的小分子生物合成基因簇(BGCs)进行了荟萃分析,并鉴定出两种梭状芽孢杆菌衍生的BGCs与主要IBD类型克罗恩病(CD)显著相关。利用合成生物学,我们发现并解决了六种脂肪酸酰胺的结构,作为富含cd的bgc的产物,我们随后在IBD患者的粪便样本中检测到它们。最后,我们表明发现的分子破坏肠道通透性,并加剧化学或遗传易感结肠炎小鼠模型的疾病。这些发现表明,微生物组衍生的小分子可能在IBD的病因学中发挥作用,并代表了发现疾病相关微生物组与宿主相互作用的分子介质的一种可推广的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


