FakET: Simulating cryo-electron tomograms with neural style transfer
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
In cryo-electron microscopy, accurate particle localization and classification are imperative. Recent deep learning solutions, though successful, require extensive training datasets. The protracted generation time of physics-based models, often employed to produce these datasets, limits their broad applicability. We introduce FakET, a method based on neural style transfer, capable of simulating the forward operator of any cryo transmission electron microscope. It can be used to adapt a synthetic training dataset according to reference data producing high-quality simulated micrographs or tilt-series. To assess the quality of our generated data, we used it to train a state-of-the-art localization and classification architecture and compared its performance with a counterpart trained on benchmark data. Remarkably, our technique matches the performance, boosts data generation speed , uses less memory, and scales well to typical transmission electron microscope detector sizes. It leverages GPU acceleration and parallel processing. The source code is available at https://github.com/paloha/faket/.
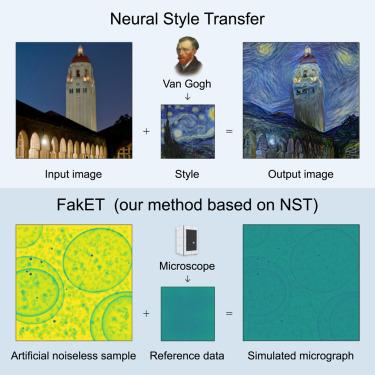
FakET:用神经风格转移模拟低温电子断层图
在低温电子显微镜中,精确的粒子定位和分类是必不可少的。最近的深度学习解决方案虽然成功,但需要大量的训练数据集。通常用于生成这些数据集的基于物理的模型的生成时间较长,限制了它们的广泛适用性。本文介绍了一种基于神经风格迁移的FakET方法,该方法能够模拟任何低温透射电子显微镜的正演算子。它可用于根据参考数据调整合成训练数据集,产生高质量的模拟显微图或倾斜序列。为了评估生成数据的质量,我们使用它来训练最先进的定位和分类体系结构,并将其性能与在基准数据上训练的对应体系结构进行比较。值得注意的是,我们的技术匹配了性能,提高了数据生成速度750×750×,使用33×33×更少的内存,并且可以很好地扩展到典型的透射电子显微镜探测器尺寸。它利用GPU加速和并行处理。源代码可从https://github.com/paloha/faket/获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


