Histone H3 lysine 4 methylation recruits DNA demethylases to enforce gene expression in Arabidopsis
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
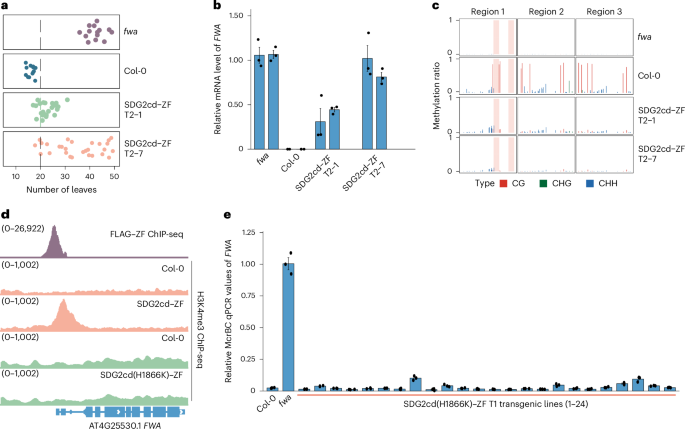
Patterning of DNA methylation in eukaryotic genomes is controlled by de novo methylation, maintenance mechanisms and demethylation pathways. In Arabidopsis thaliana, DNA demethylation enzymes are clearly important for shaping methylation patterns, but how they are regulated is poorly understood. Here we show that the targeting of histone H3 lysine four trimethylation (H3K4me3) with the catalytic domain of the SDG2 histone methyltransferase potently erased DNA methylation and gene silencing at FWA and also erased CG DNA methylation in many other regions of the Arabidopsis genome. This methylation erasure was completely blocked in the ros1 dml2 dml3 triple mutant lacking DNA demethylation enzymes, showing that H3K4me3 promotes the active removal of DNA methylation. Conversely, we found that the targeted removal of H3K4me3 increased the efficiency of targeted DNA methylation. These results highlight H3K4me3 as a potent anti-DNA methylation mark and also pave the way for development of more powerful epigenome engineering tools. This study revealed that targeting H3K4me3 via the H3K4 methyltransferase SDG2 activates gene expression and removes DNA methylation by recruiting DNA demethylases. Conversely, the removal of H3K4me3 synergistically enhances targeted DNA methylation.

拟南芥中组蛋白H3赖氨酸4甲基化招募DNA去甲基化酶来加强基因表达
真核生物基因组中DNA甲基化的模式受新生甲基化、维持机制和去甲基化途径的控制。在拟南芥中,DNA去甲基化酶显然对甲基化模式的形成很重要,但它们是如何被调节的却知之甚少。在这里,我们发现用SDG2组蛋白甲基转移酶的催化结构域靶向组蛋白H3赖氨酸4三甲基化(H3K4me3)可以有效地消除FWA的DNA甲基化和基因沉默,也可以消除拟南芥基因组许多其他区域的CG DNA甲基化。在缺乏DNA去甲基化酶的ros1 dml2 dml3三重突变体中,这种甲基化消除被完全阻断,表明H3K4me3促进了DNA甲基化的主动去除。相反,我们发现靶向去除H3K4me3可以提高靶向DNA甲基化的效率。这些结果突出了H3K4me3作为一种有效的抗dna甲基化标记,也为开发更强大的表观基因组工程工具铺平了道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


