Dynamic integration of feature- and template-based methods improves the prediction of conformational B cell epitopes
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The accurate prediction of conformational epitopes promotes our understanding of antigen-antibody interactions. All existing algorithms depend on a feature-based strategy, which limits their performance. A template-based strategy can provide complementary information, and the interplay between these two strategies could improve the prediction of epitopes. Here, we present DynaBCE, a dynamic ensemble algorithm to effectively identify conformational B cell epitopes (BCEs). Using novel handcrafted structural descriptors and embeddings from protein language models, we developed machine learning and deep learning modules based on boosting algorithms and geometric graph neural networks, respectively. Furthermore, we built a template module by leveraging known structural template information and transformer-based algorithms to capture binding signatures. Finally, we integrated the three modules using a dynamic weighting approach to maximize the strength of each module for different samples. DynaBCE achieved promising results for both native and predicted structures and outperformed previous methods as demonstrated in various evaluation scenarios.
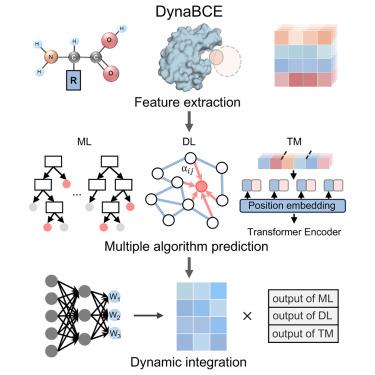
基于特征和模板的方法的动态集成提高了构象B细胞表位的预测
构象表位的准确预测促进了我们对抗原-抗体相互作用的理解。所有现有的算法都依赖于基于特征的策略,这限制了它们的性能。基于模板的策略可以提供互补的信息,这两种策略之间的相互作用可以提高表位的预测。在这里,我们提出DynaBCE,一个动态集成算法,有效地识别构象B细胞表位(bce)。利用新的手工结构描述符和蛋白质语言模型的嵌入,我们分别基于增强算法和几何图神经网络开发了机器学习和深度学习模块。此外,我们利用已知的结构模板信息和基于转换器的算法构建了模板模块来捕获绑定签名。最后,我们使用动态加权方法整合三个模块,以最大限度地提高每个模块对不同样本的强度。DynaBCE在原生结构和预测结构上都取得了令人鼓舞的结果,并在各种评估场景中优于以前的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


