Development and Validation of Prognostic Characteristics Associated With Chromatin Remodeling-Related Genes in Ovarian Cancer
Abstract
Background
Ovarian cancer (OC) is a prevalent malignant tumor in the field of gynecology, exhibiting the third highest incidence rate and the highest mortality rate among gynecological tumors. Chromatin remodeling accomplishes specific chromatin condensation at distinct genomic loci and plays an essential role in epigenetic regulation associated with various processes related to cancer development.
Methods
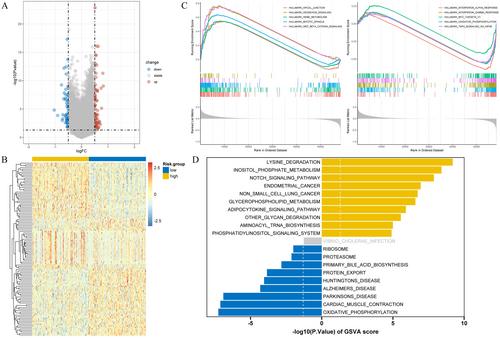
Differentially expressed genes (DEGs) between OC and control samples were screened from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases, combined with chromatin remodeling-related genes (CRRGs) obtained from the GeneCards database to identify differentially expressed CRRGs (DECRRGs). Enrichment analysis and protein–protein interaction (PPI) network were performed on the DECRRGs. Prognostic genes of OC were screened using univariate Cox and least absolute shrinkage and selection operator (Lasso) analyses. A risk model based on prognostic genes was developed, and the survival probability of OC patients in different risk groups was analyzed by Kaplan–Meier (KM) curve. Finally, the expression levels of prognostic genes were validated by quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting.
Results
In total, 7 potential prognostic genes associated with the progression of OC patients were obtained, including ARID1B, ATRX, CHRAC1, HDAC1, INO80, MBD2, and SS18. Based on the expression level of prognostic genes, OC patients were divided into high-risk group and low-risk group. Survival analysis indicated that patients classified into the high-risk group had higher mortality rates, which enables this prediction model to be utilized as an independent predictor of OC. Immunocorrelation analysis showed that low-risk patients were more likely to benefit from immunotherapy.
Conclusion
In this study, we have identified 7 prognostic genes, including ARID1B, ATRX, CHRAC1, HDAC1, INO80, MBD2, and SS18. Overall, our findings provided a foundation for further comprehension of the potential molecular mechanisms underlying OC pathogenesis and progression.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


