Remote Regulation by VirB, the Transcriptional Anti‐Silencer of Shigella Virulence Genes, Provides Mechanistic Information
IF 2.6
2区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
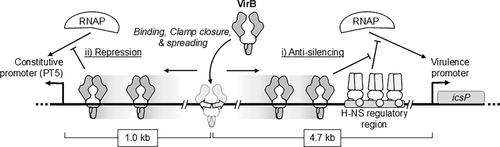
Classical models of bacterial transcription show regulators binding close to promoter elements to exert their effect. However, the scope for long‐range regulation exists, especially by nucleoid structuring proteins, like H‐NS. Here, long‐range regulation by VirB, a transcriptional regulator that alleviates H‐NS‐mediated silencing of key virulence genes in
志贺氏菌毒力基因转录抗沉默者VirB的远程调控提供了机制信息
经典的细菌转录模型显示,调节因子结合在启动子元件附近以发挥其作用。然而,长期调控的范围是存在的,特别是通过类核结构蛋白,如H‐NS。本文在体内研究了VirB(一种转录调节因子,可缓解H - NS介导的志贺氏菌中关键毒力基因的沉默)的远程调控,以测试远程调控的局限性,并提供进一步的机制见解。如果icsP启动子的同源位点在质粒报告子中比其原生位置上游重新定位1kb、3.3 kb,甚至4.7 kb,那么其依赖VirB的调控仍然存在。依赖VirB的调控随着结合位点距离的增加而减弱。虽然细胞VirB池的增加提高了所有具有野生型VirB结合位点的构建体的启动子活性,但与天然位点相比,远程位点的启动子活性并没有不成比例的增加。由于VirB在停靠在其- 35元件附近时阻断了一个组成活性启动子(PT5),因此我们下一步将VirB的结合位点移到了启动子区域之外。我们发现VirB仍然干扰启动子的活性。这些发现以及来自VirB结合位点周围的分子障碍的发现与VirB转录调控的各种模型相一致。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Microbiology
生物-生化与分子生物学
CiteScore
7.20
自引率
5.60%
发文量
132
审稿时长
1.7 months
期刊介绍:
Molecular Microbiology, the leading primary journal in the microbial sciences, publishes molecular studies of Bacteria, Archaea, eukaryotic microorganisms, and their viruses.
Research papers should lead to a deeper understanding of the molecular principles underlying basic physiological processes or mechanisms. Appropriate topics include gene expression and regulation, pathogenicity and virulence, physiology and metabolism, synthesis of macromolecules (proteins, nucleic acids, lipids, polysaccharides, etc), cell biology and subcellular organization, membrane biogenesis and function, traffic and transport, cell-cell communication and signalling pathways, evolution and gene transfer. Articles focused on host responses (cellular or immunological) to pathogens or on microbial ecology should be directed to our sister journals Cellular Microbiology and Environmental Microbiology, respectively.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


