Multi-interface licensing of protein import into a phage nucleus
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
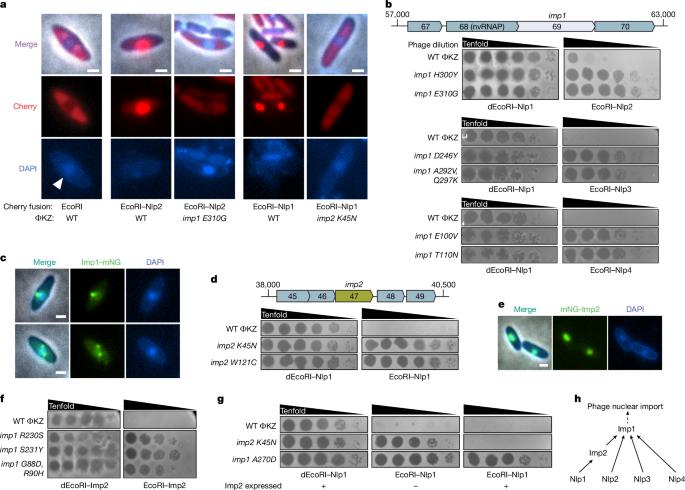
Bacteriophages use diverse mechanisms to evade antiphage defence systems. ΦKZ-like jumbo phages assemble a proteinaceous, nucleus-like compartment that excludes antagonistic host nucleases and also internalizes DNA replication and transcription machinery1–4. The phage factors required for protein import and the mechanisms of selectivity remain unknown, however. Here we uncover an import system comprising proteins highly conserved across nucleus-forming phages, together with additional cargo-specific contributors. Using a genetic selection that forces the phage to decrease or abolish the import of specific proteins, we determine that the importation of five different phage nuclear-localized proteins requires distinct interfaces of the same factor, Imp1 (gp69). Imp1 localizes early to the nascent phage nucleus and forms discrete puncta in the mature phage nuclear periphery, probably in complex with direct interactor Imp6 (gp67), a conserved protein encoded in the same locus. The import of certain proteins, including a host topoisomerase, additionally requires Imp3 (gp59), a conserved factor necessary for proper Imp1 function. Three additional non-conserved phage proteins (Imp2 and Imp4/Imp5) are required for the import of two queried nuclear cargos (nuclear-localized protein 1 and host topoisomerase, respectively), perhaps acting as specific adaptors. We therefore propose a core import system that includes Imp1, Imp3 and Imp6, with multiple interfaces of Imp1 licensing transport through a protein lattice. This study uncovers a highly conserved jumbo phage protein, Imp1, that possesses multiple interfaces to license protein import into a proteinaceous nucleus-like compartment, using a genetic selection that forces the phage to decrease or abolish the import of specific proteins.
蛋白质输入噬菌体细胞核的多界面许可
噬菌体使用多种机制来逃避抗噬菌体防御系统。ΦKZ-like巨型噬菌体组装一个蛋白质,核样隔室,排除拮抗宿主核酸酶,也内化DNA复制和转录机制1,2,3,4。然而,蛋白质输入所需的噬菌体因子和选择性机制仍不清楚。在这里,我们发现了一个进口系统,包括在核形成噬菌体中高度保守的蛋白质,以及额外的货物特异性贡献者。利用基因选择迫使噬菌体减少或取消特定蛋白质的输入,我们确定五种不同的噬菌体核定位蛋白的输入需要同一因子Imp1 (gp69)的不同界面。Imp1定位于早期的噬菌体细胞核,并在成熟的噬菌体核周围形成离散的点,可能与直接相互作用物Imp6 (gp67)复杂,后者是编码在同一位点的保守蛋白。某些蛋白质的输入,包括宿主拓扑异构酶,还需要Imp3 (gp59),这是一个保守因子,是Imp1正常功能所必需的。另外三种非保守噬菌体蛋白(Imp2和Imp4/Imp5)需要用于两个查询核货物(分别为核定位蛋白1和宿主拓扑异构酶)的进口,可能作为特定的适配器。因此,我们提出了一个包含Imp1、Imp3和Imp6的核心进口系统,其中Imp1的多个接口允许通过蛋白质晶格进行运输。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


