Overlap of Genomic and Transcriptomic Genes Identified in Familial Eosinophilic Esophagitis
IF 25.7
1区 医学
Q1 GASTROENTEROLOGY & HEPATOLOGY
引用次数: 0
Abstract
Background and Aims
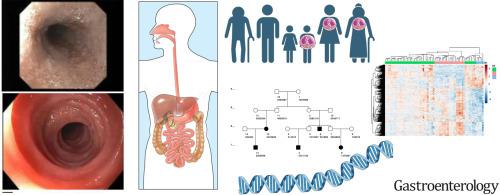
Evidence for a genetic contribution to eosinophilic esophagitis (EoE) exists from family and genome-wide association studies. Extensive investigation into rare variants contributing to EoE has not been performed. The study aim is to evaluate families with multiple cases of EoE by genomic and transcriptomic sequencing to identify genes predisposing to EoE.Methods
We whole exome sequenced (WES) distant relative pairs (e.g., cousins) in extended EoE families and other affected relatives to identify rare, shared, potentially pathologic variants. RNA-Seq was performed in nuclear families with multiple EoE cases. We compared the overlap of genes from DNA and RNA sequencing for relevance to disease manifestations.Results
WES was performed in 50 familial cases in 21 EoE extended pedigrees. We observed 189 rare, candidate predisposition variants in 181 genes with complete sharing among all affected family members within each pedigree. RNA-Seq was performed for 43 EoE cases in 18 nuclear families, including 6 relatives without EoE. We observed 698 total differentially expressed genes compared to controls. We identified three genes (MUC16, ADGRE1, and TENM3) with evidence of rare variant sharing among all affected family members and differential gene expression. We identified 36 other genes with partial sharing of rare variants among some affected family members and with differential gene expression. Several genes identified as prominent in EoE were also differentially expressed in unaffected relatives.Conclusions
Genes related to immune response, barrier dysfunction, and cell adhesion were identified in familial EoE cases and unaffected family members, supporting a genetic familial predisposition and a possible multi-hit background to disease pathophysiology.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Gastroenterology
医学-胃肠肝病学
CiteScore
45.60
自引率
2.40%
发文量
4366
审稿时长
26 days
期刊介绍:
Gastroenterology is the most prominent journal in the field of gastrointestinal disease. It is the flagship journal of the American Gastroenterological Association and delivers authoritative coverage of clinical, translational, and basic studies of all aspects of the digestive system, including the liver and pancreas, as well as nutrition.
Some regular features of Gastroenterology include original research studies by leading authorities, comprehensive reviews and perspectives on important topics in adult and pediatric gastroenterology and hepatology. The journal also includes features such as editorials, correspondence, and commentaries, as well as special sections like "Mentoring, Education and Training Corner," "Diversity, Equity and Inclusion in GI," "Gastro Digest," "Gastro Curbside Consult," and "Gastro Grand Rounds."
Gastroenterology also provides digital media materials such as videos and "GI Rapid Reel" animations. It is abstracted and indexed in various databases including Scopus, Biological Abstracts, Current Contents, Embase, Nutrition Abstracts, Chemical Abstracts, Current Awareness in Biological Sciences, PubMed/Medline, and the Science Citation Index.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


