The structure of a NEMO construct engineered for screening reveals novel determinants of inhibition
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
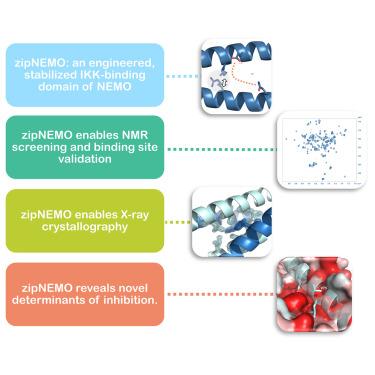
NEMO is an essential component in the activation of the canonical nuclear factor κB (NF-κB) pathway and exerts its function by recruiting the IκB kinases (IKK) to the IKK complex. Inhibition of the NEMO/IKKs interaction is an attractive therapeutic paradigm for diseases related to NF-κB mis-regulation, but a difficult endeavor because of the extensive protein-protein interface. Here we report the design and characterization of novel engineered constructs of the IKK-binding domain of NEMO, programmed to render this difficult protein domain amenable to NMR measurements and crystallization, while preserving its biological function. ZipNEMO binds IKKβ with nanomolar affinity, is amenable to heteronuclear nuclear magnetic resonance (NMR) techniques and structure determination by X-ray crystallography. We show that NMR spectra of zipNEMO allow to detect inhibitor binding in solution and resonance assignment. The crystal structure of zipNEMO reveals a novel ligand binding motif and the adaptability of the binding pocket and inspired the design of new peptide inhibitors.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


