A human gut bacterium antagonizes neighboring bacteria by altering their protein-folding ability
IF 20.6
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
Antagonistic interactions play a key role in determining microbial community dynamics. Here, we report that one of the most widespread contact-dependent effectors in human gut microbiomes, Bte1, directly targets the PpiD-YfgM periplasmic chaperone complex in related microbes. Structural, biochemical, and genetic characterization of this interaction reveals that Bte1 reverses the activity of the chaperone complex, promoting substrate aggregation and toxicity. Using Bacteroides, we show that Bte1 is active in the mammalian gut, conferring a fitness advantage to expressing strains. Recipient cells targeted by Bte1 exhibit sensitivity to membrane-compromising conditions, and human gut microbes can use this effector to exploit pathogen-induced inflammation in the gut. Further, Bte1 allelic variation in gut metagenomes provides evidence for an arms race between Bte1-encoding and immunity-encoding strains in humans. Together, these studies demonstrate that human gut microbes alter the protein-folding capacity of neighboring cells and suggest strategies for manipulating community dynamics.
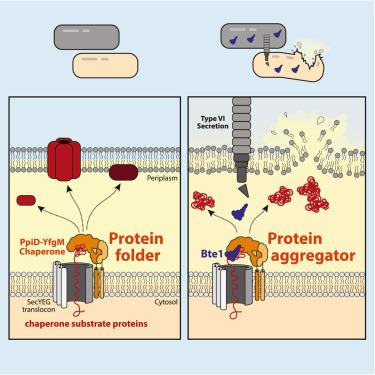
人类肠道细菌通过改变邻近细菌的蛋白质折叠能力来对抗它们
拮抗相互作用在决定微生物群落动态中起关键作用。在这里,我们报道了人类肠道微生物组中最广泛的接触依赖效应物之一Bte1直接靶向相关微生物中的PpiD-YfgM质周伴侣复合物。这种相互作用的结构、生化和遗传特征表明,Bte1逆转伴侣复合物的活性,促进底物聚集和毒性。利用拟杆菌,我们发现Bte1在哺乳动物肠道中是活跃的,赋予表达菌株适应性优势。Bte1靶向的受体细胞表现出对膜损害条件的敏感性,人类肠道微生物可以利用这种效应物在肠道中利用病原体诱导的炎症。此外,肠道宏基因组中的Bte1等位基因变异为人类Bte1编码菌株和免疫编码菌株之间的军备竞赛提供了证据。总之,这些研究表明,人类肠道微生物改变了邻近细胞的蛋白质折叠能力,并提出了操纵群落动态的策略。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


