Interplay between gut microbial communities and metabolites modulates pan-cancer immunotherapy responses
IF 27.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Immune checkpoint blockade (ICB) therapy has revolutionized cancer treatment but remains effective in only a subset of patients. Emerging evidence suggests that the gut microbiome and its metabolites critically influence ICB efficacy. In this study, we performed a multi-omics analysis of fecal microbiomes and metabolomes from 165 patients undergoing anti-programmed cell death protein 1 (PD-1)/programmed death ligand 1 (PD-L1) therapy, identifying microbial and metabolic entities associated with treatment response. Integration of data from four public metagenomic datasets (n = 568) uncovered cross-cohort microbial and metabolic signatures, validated in an independent cohort (n = 138). An integrated predictive model incorporating these features demonstrated robust performance. Notably, we characterized five response-associated enterotypes, each linked to specific bacterial taxa and metabolites. Among these, the metabolite phenylacetylglutamine (PAGln) was negatively correlated with response and shown to attenuate anti-PD-1 efficacy in vivo. This study sheds light on the interplay among the gut microbiome, the gut metabolome, and immunotherapy response, identifying potential biomarkers to improve treatment outcomes.
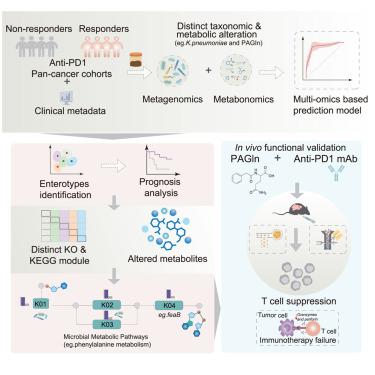
肠道微生物群落和代谢物之间的相互作用调节泛癌症免疫治疗反应
免疫检查点阻断(ICB)疗法已经彻底改变了癌症治疗,但仍然只对一小部分患者有效。新出现的证据表明,肠道微生物群及其代谢物对ICB的疗效有关键影响。在这项研究中,我们对165名接受抗程序性细胞死亡蛋白1 (PD-1)/程序性死亡配体1 (PD-L1)治疗的患者的粪便微生物组和代谢组进行了多组学分析,确定了与治疗反应相关的微生物和代谢实体。整合来自四个公共宏基因组数据集(n = 568)的数据揭示了跨队列微生物和代谢特征,并在一个独立队列(n = 138)中得到验证。结合这些特征的集成预测模型显示了稳健的性能。值得注意的是,我们描述了五种与反应相关的肠型,每一种都与特定的细菌分类群和代谢物有关。其中,代谢物苯乙酰谷氨酰胺(PAGln)与反应呈负相关,并在体内减弱抗pd -1的疗效。这项研究揭示了肠道微生物组、肠道代谢组和免疫治疗反应之间的相互作用,确定了改善治疗结果的潜在生物标志物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Cell metabolism
生物-内分泌学与代谢
CiteScore
48.60
自引率
1.40%
发文量
173
审稿时长
2.5 months
期刊介绍:
Cell Metabolism is a top research journal established in 2005 that focuses on publishing original and impactful papers in the field of metabolic research.It covers a wide range of topics including diabetes, obesity, cardiovascular biology, aging and stress responses, circadian biology, and many others.
Cell Metabolism aims to contribute to the advancement of metabolic research by providing a platform for the publication and dissemination of high-quality research and thought-provoking articles.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


