Architecture of an embracing lipase-foldase complex of the type II secretion system of Acinetobacter baumannii
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
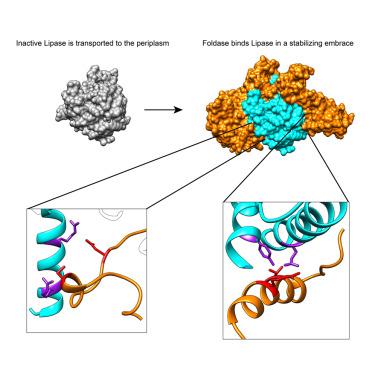
Acinetobacter baumannii is a major human pathogen responsible for a growing number of multi-antibiotic-resistant infections, and of critical priority for the World Health Organization (WHO). A. baumannii employs a type II secretion system (T2SS) to secrete toxins extracellularly to enable cytotoxicity and colonization. Lipase LipA, secreted by the A. baumannii T2SS, is required for virulence and fitness, and in the periplasm is maintained in an active state by its essential foldase, LipB. Here we report that LipA is able to recognize lipids of different chain lengths at extremes of pH and temperature, thanks to its stabilization by LipB through an extended, highly helical “embrace.” A vast bioinformatic analysis indicates that LipB-like foldases are widespread over numerous proteobacteria, and thus the extended foldase architecture shown here could be widespread. These results provide new insight into A. baumannii’s adaptability as a pathogen in different environments and could facilitate the development of novel antibacterials.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


