SAMPL-seq reveals micron-scale spatial hubs in the human gut microbiome
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
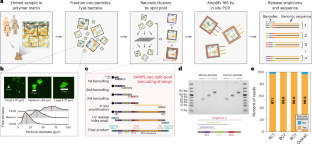
The local arrangement of microbes can profoundly impact community assembly, function and stability. However, our understanding of the spatial organization of the human gut microbiome at the micron scale is limited. Here we describe a high-throughput and streamlined method called Split-And-pool Metagenomic Plot-sampling sequencing (SAMPL-seq) to capture spatial co-localization in a complex microbial consortium. The method obtains microbial composition of micron-scale subcommunities through split-and-pool barcoding. SAMPL-seq analysis of the healthy human gut microbiome identified bacterial taxa pairs that consistently co-occurred both over time and across multiple individuals. These co-localized microbes organize into spatially distinct groups or ‘spatial hubs’ dominated by Bacteroidaceae, Ruminococcaceae and Lachnospiraceae families. Using inulin as a dietary perturbation, we observed reversible spatial rearrangement of the gut microbiome where specific taxa form new local partnerships. Spatial metagenomics using SAMPL-seq can unlock insights into microbiomes at the micron scale. Split-And-pool Metagenomic Plot-sampling sequencing (SAMPL-seq) can be applied to complex microbial communities to reveal spatial co-localization of microbes at the micron scale.

SAMPL-seq揭示了人类肠道微生物组中微米尺度的空间枢纽
微生物的局部排列对群落的组装、功能和稳定性有着深远的影响。然而,我们对人类肠道微生物群在微米尺度上的空间组织的理解是有限的。在这里,我们描述了一种高通量和流线化的方法,称为分裂-池宏基因组图谱采样测序(SAMPL-seq),以捕获复杂微生物群体中的空间共定位。该方法通过split-and-pool条形码获取微米尺度亚群落的微生物组成。健康人类肠道微生物组的SAMPL-seq分析鉴定出随时间和在多个个体中一致共同发生的细菌分类群对。这些共定位的微生物在空间上形成不同的群体或“空间中心”,以拟杆菌科、瘤胃球菌科和毛菌科为主。使用菊粉作为膳食扰动,我们观察到肠道微生物组的可逆空间重排,其中特定分类群形成新的局部伙伴关系。使用SAMPL-seq的空间宏基因组学可以在微米尺度上解锁对微生物组的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


