Genome-wide gene expression profiles throughout human malaria parasite liver stage development in humanized mice
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
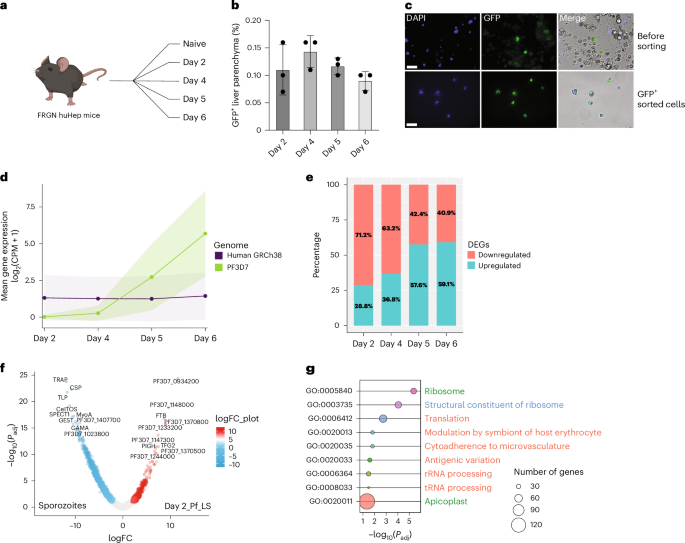
Gene expression of Plasmodium falciparum (Pf) liver-stage (LS) parasites has remained poorly characterized, although they are major vaccine and drug targets. Using a human liver-chimaeric mouse model and a fluorescent parasite line (PfNF54CSPGFP), we isolated PfLS and performed transcriptomics on key LS developmental phases. We linked clustered gene expression to ApiAP2, a major family of transcription factors that regulate the parasite life cycle. This provided insights into transcriptional regulation of LS infection and expression of essential LS metabolic and biosynthetic pathways. We observed expression of antigenically variant PfEMP1 proteins and the major Pf protein export machine PTEX and identified protein candidates that might be exported by LS parasites. Comparing Pf and P. vivax LS transcriptomes, we uncovered differences in their expression of sexual commitment factors. This data will aid LS research and vaccine and drug target identification for prevention of malaria infection. Transcriptomics of gene expression in Plasmodium falciparum liver-stage parasites reveals transcriptional regulation, metabolic pathways and antigen expression, facilitating vaccine and drug target identification.

全基因组基因表达谱在人源化小鼠中贯穿人疟疾寄生虫肝脏发育阶段
恶性疟原虫(Pf)肝期(LS)寄生虫的基因表达特征仍然很差,尽管它们是主要的疫苗和药物靶点。利用人肝脏嵌合小鼠模型和荧光寄生虫系(PfNF54CSPGFP),我们分离了PfLS,并对LS的关键发育阶段进行了转录组学研究。我们将集群基因表达与ApiAP2联系起来,ApiAP2是调节寄生虫生命周期的主要转录因子家族。这为LS感染的转录调控以及LS基本代谢和生物合成途径的表达提供了见解。我们观察了抗原性变异的PfEMP1蛋白和主要的Pf蛋白输出机器PTEX的表达,并鉴定了可能由LS寄生虫输出的候选蛋白。比较Pf和间日疟原虫LS转录组,我们发现了它们在性承诺因子表达上的差异。这些数据将有助于LS研究以及预防疟疾感染的疫苗和药物靶点的确定。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


